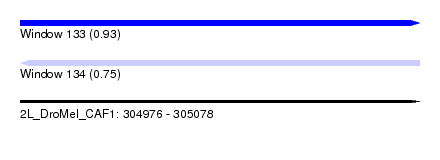
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 304,976 – 305,078 |
| Length | 102 |
| Max. P | 0.928439 |

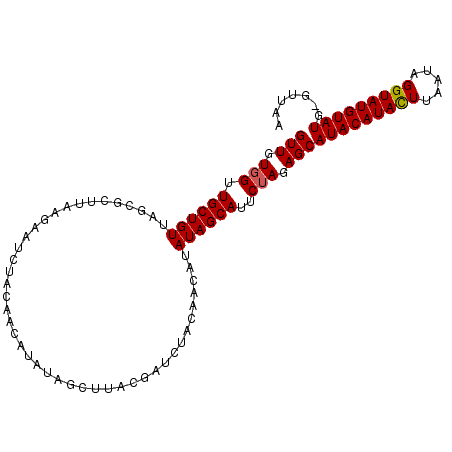
| Location | 304,976 – 305,078 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 85.37 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -14.64 |
| Energy contribution | -17.95 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
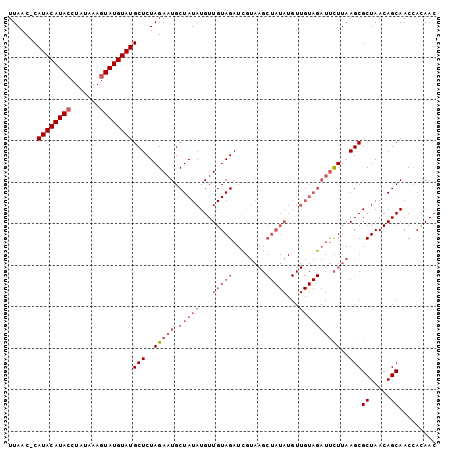
>2L_DroMel_CAF1 304976 102 + 22407834 UUAAC-CAUACAUACCUAUAAAGUAUGUAUGCUCUAGAAUGCUAUAUGUUGUAGAUCGUAAGCUAUAUGUUGUAGAUUCUUAAGCGCUAACAGCAACCACAAC .....-.((((((((.......))))))))(((..(((((.(((((...(((((..(....))))))...))))))))))..)))((.....))......... ( -24.60) >DroSim_CAF1 9586 102 + 1 UUAAC-CAUACAUACCUAUUAAGUAUGUAUGCUCUAGAAUGCUAUAUGUUGUAGAUCGUAAGCUAUAUGUUGUAGAUUCUUAAGCGCUAACAGCAACCACAAC .....-.((((((((.......))))))))(((..(((((.(((((...(((((..(....))))))...))))))))))..)))((.....))......... ( -24.60) >DroYak_CAF1 9885 102 + 1 UUAAC-CAUACAUACCUAUUAAGUAUGUAUGCUCUAGAAUGCUAUAUGUUGUAGAUUGUAAGCUAUAUGUUGUAGAUUCUUAAGCGCUAACAGCAACCAUAAC .....-.((((((((.......))))))))(((..(((((.(((((...(((((........)))))...))))))))))..)))((.....))......... ( -24.50) >DroAna_CAF1 12533 82 + 1 UUAACCCAUACAUACCUAUAUAAUAUGUAUGCUCUAGAAUGCUAUAUGUUGUAGAU---------------------CUUUAAGCGCUAACAGCAACAACAAC ..................(((((((((((.((........)))))))))))))...---------------------......((.......))......... ( -12.10) >consensus UUAAC_CAUACAUACCUAUAAAGUAUGUAUGCUCUAGAAUGCUAUAUGUUGUAGAUCGUAAGCUAUAUGUUGUAGAUUCUUAAGCGCUAACAGCAACCACAAC .......((((((((.......))))))))(((..(((((.(((((...(((((........)))))...))))))))))..)))((.....))......... (-14.64 = -17.95 + 3.31)

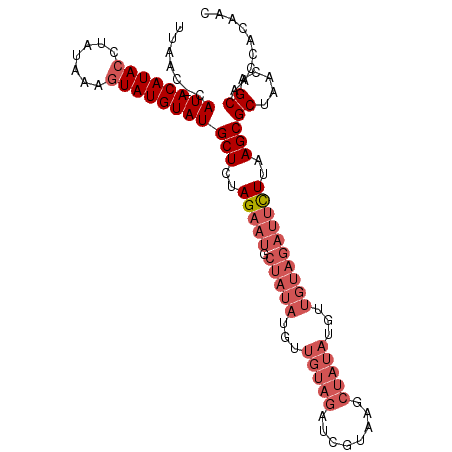
| Location | 304,976 – 305,078 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 85.37 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -15.91 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 304976 102 - 22407834 GUUGUGGUUGCUGUUAGCGCUUAAGAAUCUACAACAUAUAGCUUACGAUCUACAACAUAUAGCAUUCUAGAGCAUACAUACUUUAUAGGUAUGUAUG-GUUAA (((((((....(((.(((......................))).)))..))))))).............((.((((((((((.....))))))))))-.)).. ( -24.65) >DroSim_CAF1 9586 102 - 1 GUUGUGGUUGCUGUUAGCGCUUAAGAAUCUACAACAUAUAGCUUACGAUCUACAACAUAUAGCAUUCUAGAGCAUACAUACUUAAUAGGUAUGUAUG-GUUAA (((((((....(((.(((......................))).)))..))))))).............((.((((((((((.....))))))))))-.)).. ( -24.65) >DroYak_CAF1 9885 102 - 1 GUUAUGGUUGCUGUUAGCGCUUAAGAAUCUACAACAUAUAGCUUACAAUCUACAACAUAUAGCAUUCUAGAGCAUACAUACUUAAUAGGUAUGUAUG-GUUAA ((((((((((.(((.(((......................))).))).....)))).))))))......((.((((((((((.....))))))))))-.)).. ( -23.15) >DroAna_CAF1 12533 82 - 1 GUUGUUGUUGCUGUUAGCGCUUAAAG---------------------AUCUACAACAUAUAGCAUUCUAGAGCAUACAUAUUAUAUAGGUAUGUAUGGGUUAA ((((((((((..(....(.......)---------------------..)..))))).)))))....(((..((((((((((.....))))))))))..))). ( -16.20) >consensus GUUGUGGUUGCUGUUAGCGCUUAAGAAUCUACAACAUAUAGCUUACGAUCUACAACAUAUAGCAUUCUAGAGCAUACAUACUUAAUAGGUAUGUAUG_GUUAA (((.(((.((((((............................................))))))..))).)))(((((((((.....)))))))))....... (-15.91 = -15.97 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:27 2006