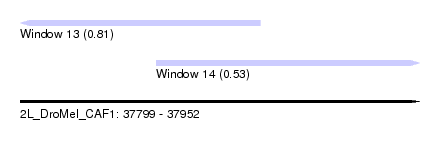
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 37,799 – 37,952 |
| Length | 153 |
| Max. P | 0.812564 |

| Location | 37,799 – 37,891 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.92 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -16.36 |
| Energy contribution | -17.17 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
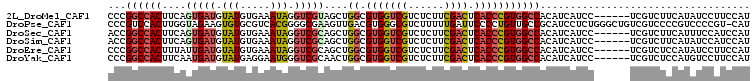
>2L_DroMel_CAF1 37799 92 - 22407834 AGCUACGACCUAUUUCACAUACAUCACUGAAGUGGCCGGGACAAAACUACCGCGCCAUUGACGCACCCCCUCUUCCACAGGUUUCUCAGCA------A .(((..(((((...(((..........)))(((((((((..........))).))))))...................)))))....))).------. ( -18.90) >DroSec_CAF1 15762 92 - 1 AGCUGCGACCUAUUUCACAUACAUCACUGAAGUGGCCGGUACAAAACUACCGCGCCAUUGACUCACCCCCCCUCCCGCAGGUUUCUCAGCA------A .((((.(((((...(((..........)))(((((((((((......))))).))))))...................)))))...)))).------. ( -24.40) >DroSim_CAF1 17342 92 - 1 AGCUGCGACCUAUUUCACAUACAUCACUGAAGUGGCCGGUACAAAACUACCGCGCCAUUGACUCACCCCCCCUCCCGCAGGUUUCUCAGCA------A .((((.(((((...(((..........)))(((((((((((......))))).))))))...................)))))...)))).------. ( -24.40) >DroEre_CAF1 15891 92 - 1 AGCUGCGACCUAUUUCACAUACAUCAAUAAAGUGGCCGGGCCAAAACUACCGUGCCAUUGACGCACACCCUUCCGCGCAGGUUCCUCAGCA------A .((((.(((((...................(((((((((..........))).))))))(.(((..........)))))))))...)))).------. ( -25.40) >DroYak_CAF1 14593 92 - 1 AGUUGCGACCCAUUCCUCAUACAUCAUUGAAGUGGCCGGGCCAAAACUACCGCGCCAUUGACGCACCCCCUUCCCCACCGAUUUCUCAGCA------A ...((((.........(((........)))(((((((((..........))).))))))..))))..........................------. ( -17.90) >DroAna_CAF1 14802 98 - 1 AGCUGCGACCUGUGGGGCAGCGCUCCUCCAAGUGGCCGGGGCAGAACUACCGUGCCAUUGACGCUCCUCCGCCACCCCAGAAUUUUCAGCAGCAACAG .(((((.....((((((.((((.((.....(((((((((..........))).)))))))))))).)))))).......((....)).)))))..... ( -35.80) >consensus AGCUGCGACCUAUUUCACAUACAUCACUGAAGUGGCCGGGACAAAACUACCGCGCCAUUGACGCACCCCCUCCCCCGCAGGUUUCUCAGCA______A .((((.(((((..........((....)).(((((((((..........))).))))))...................)))))...))))........ (-16.36 = -17.17 + 0.81)

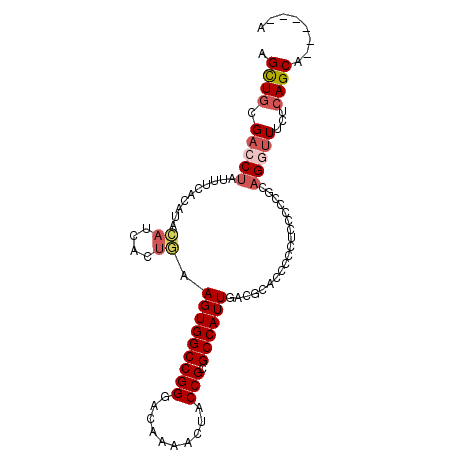
| Location | 37,851 – 37,952 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 83.82 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -22.62 |
| Energy contribution | -23.15 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 37851 101 + 22407834 CCCGGCCACUUCAGUGAUGUAUGUGAAAUAGGUCGUAGCUGGCGUGGUCGUCUCUUCGACUCACCCGUGGCCACAUCAUCC------UCGUCUUCAUAUCCUUCCAU ...((((((......(((.(((.....))).)))......((.(((((((......)))).))))))))))).........------.................... ( -28.70) >DroPse_CAF1 16555 106 + 1 CCCGUCCACUUGGUAGAAGUGUGCGUCACGGGGCGAAGUUGACGUGGGCGUCUUUUUGAUUCCCCUGUUGCCGCAUCCUCUGGGCUGUCGUCCCCGUCCCCGU-CAU ..((..(((((.....)))))..))..(((((((((....((((.(((.(((.....))).))).))))(((.((.....)))))..))).))))))......-... ( -36.40) >DroSec_CAF1 15814 101 + 1 ACCGGCCACUUCAGUGAUGUAUGUGAAAUAGGUCGCAGCUGGCGUGGUCGUCUCUUCGACUCACCCGUGGCCACAUCAUCC------UCGUCUUCAUUUCCAUCCAU ...((((((....(((((.(((.....))).)))))....((.(((((((......)))).))))))))))).........------.................... ( -32.80) >DroSim_CAF1 17394 101 + 1 ACCGGCCACUUCAGUGAUGUAUGUGAAAUAGGUCGCAGCUGGCGUGGUCGUCUCUUCGACUCACCCGUGGCCACAUCAUCC------UCGUCUUCAUAUCCAUCCAU ...((((((....(((((.(((.....))).)))))....((.(((((((......)))).))))))))))).........------.................... ( -32.80) >DroEre_CAF1 15943 101 + 1 CCCGGCCACUUUAUUGAUGUAUGUGAAAUAGGUCGCAGCUGGCGUGGUCGUCUCUUCGACUCACCCGUGGCCACAUCAUCC------UCGUCUCCAUAUCCUUCCAU ...((((((......(((.(((.....))).)))......((.(((((((......)))).))))))))))).........------.................... ( -28.70) >DroYak_CAF1 14645 101 + 1 CCCGGCCACUUCAAUGAUGUAUGAGGAAUGGGUCGCAACUGGCGUGGUCGUCUCUUCGACUCACCCGUGGCCACAUCAUCC------UCGUCUCCAUGUCCUUCCAU ...((........(((..(.((((((((((((((((....((.(((((((......)))).))))))))))).)))..)))------)))))..)))......)).. ( -32.54) >consensus CCCGGCCACUUCAGUGAUGUAUGUGAAAUAGGUCGCAGCUGGCGUGGUCGUCUCUUCGACUCACCCGUGGCCACAUCAUCC______UCGUCUCCAUAUCCUUCCAU ...((((((....(((((.(((.....))).)))))....((.(((((((......)))).)))))))))))................................... (-22.62 = -23.15 + 0.53)
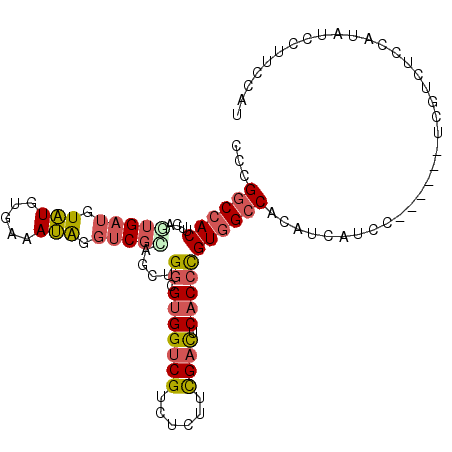
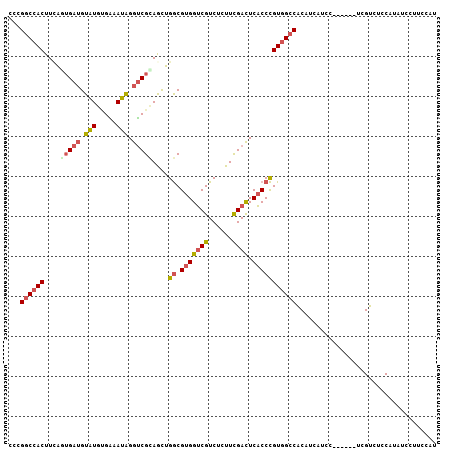
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:09 2006