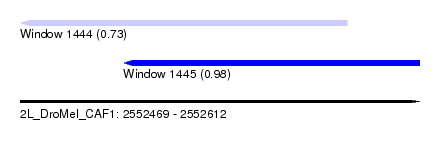
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,552,469 – 2,552,612 |
| Length | 143 |
| Max. P | 0.976867 |

| Location | 2,552,469 – 2,552,586 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
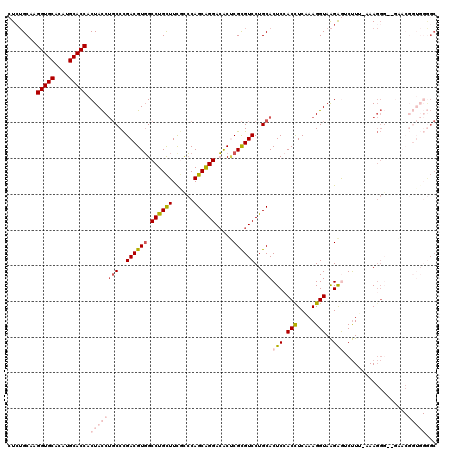
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -29.49 |
| Energy contribution | -31.30 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
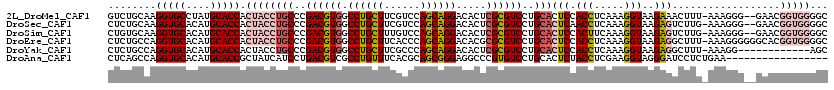
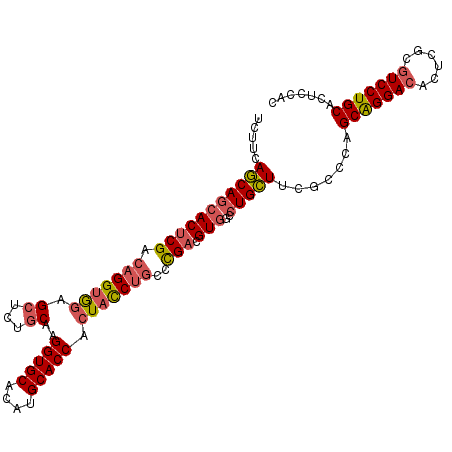
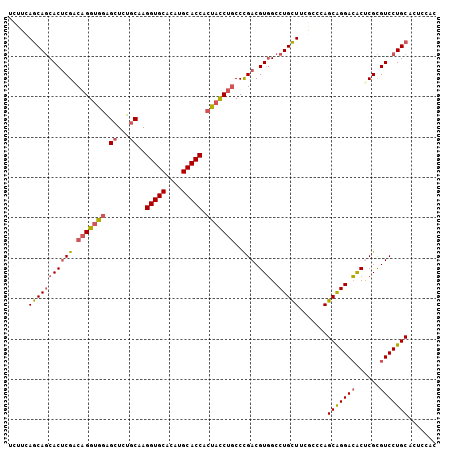
>2L_DroMel_CAF1 2552469 117 - 22407834 GUCUGCAAGGUGCCUAUGCACCACUACCUGCCCGACGUGGCCUGCUUCGUCCAGCAGGACACUCGCGUCCUGCACUCCACCUCAAAGGUAAGAAACUUU-AAAGGG--GAACGGUGGGGC ....((..(((((....)))))....((..((.((((.((....)).))))..(((((((......)))))))..(((.(((..(((((.....)))))-..))))--))..))..)))) ( -46.30) >DroSec_CAF1 131840 117 - 1 CUCUGCAAGGUGCACAUGCACCACUACCUGCCCGACGUGGCCUGCUUCGUCCAGCAGGACACUCGCGUCCUGCACUCAACCUCAAAGGUAAGAGUCUUG-AAAGGG--GAACGGUGGGGC ....((..(((((....))))).(((((..(((((((.((....)).))))..(((((((......)))))))((((.(((.....)))..))))....-....))--)...))))).)) ( -46.50) >DroSim_CAF1 135082 117 - 1 CUGUGCAAGGUGCACAUGCACCACUACCUGCCCGACGUGGCCUGCUUUGUCCAGCAGGACACUCGCGUCCUGCACUCCACUUCAAAGGUAAGAGUCUUG-AAAGGG--GAACGGUGGGGC ....((..(((((....))))).((((((((..(((((((((((((......))))))....)))))))..))).(((.(((..((((......)))).-.))).)--))..))))).)) ( -43.90) >DroEre_CAF1 136139 119 - 1 CUCUGCCAGGUGCACAUGCACCACUACCUGCCCGACGUGGCCUGCUUCACCCAGCAGGACACGCGCGUCCUGCACUCCACCUCAAAGGUAAGAGGCUUU-AAAGGGGGGCACGGUGGGGC ....(((.(((((....))))).((((((((((((.((.....)).)).(((.(((((((......))))))).(((.(((.....)))..))).....-...)))))))).)))))))) ( -55.70) >DroYak_CAF1 135855 107 - 1 CUCUGCCAGGUGCACAUGCACCACUACCUGCCCGACGUGGCCUGCUUCGCCCAGCAGGACACUCGCGUCCUGCACUCCACCUCAAAGGUAAGAGGCUUU-AAAGG------------AGC (((((((.(((((....)))))......(((..(((((((((((((......))))))....)))))))..)))............))).))))((((.-....)------------))) ( -39.20) >DroAna_CAF1 185921 103 - 1 CUCAGCCAGGUGCACAUGCACCGCUAUCAUCCUGACGUCGCCUGUUUCACGCAGCGGGAGGCCCGUGUCCUGCACUCUACCUCGAAGGUAGGGAUCCUCUGAA----------------- .((((...(((((....)))))((...((((((..(((.((.((...)).)).)))..)))...)))....)).(((((((.....))))))).....)))).----------------- ( -34.10) >consensus CUCUGCAAGGUGCACAUGCACCACUACCUGCCCGACGUGGCCUGCUUCGCCCAGCAGGACACUCGCGUCCUGCACUCCACCUCAAAGGUAAGAGUCUUU_AAAGGG__GAACGGUGGGGC ........(((((....))))).((((((((..((((((.((((((......)))))).....))))))..)))(((.(((.....)))..)))..................)))))... (-29.49 = -31.30 + 1.81)
| Location | 2,552,506 – 2,552,612 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 88.18 |
| Mean single sequence MFE | -42.77 |
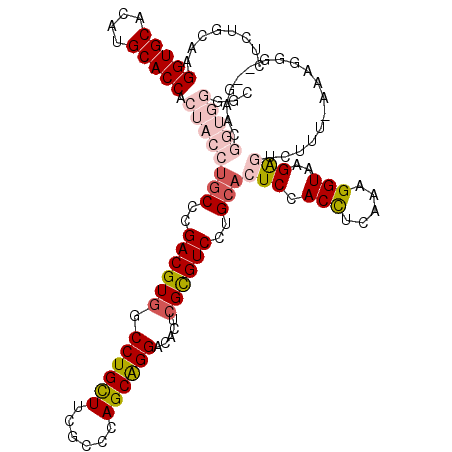
| Consensus MFE | -36.15 |
| Energy contribution | -36.82 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2552506 106 - 22407834 UCUUCAGCAGCACUCGACAGGUGCAGGUCUGCAAGGUGCCUAUGCACCACUACCUGCCCGACGUGGCCUGCUUCGUCCAGCAGGACACUCGCGUCCUGCACUCCAC ......((((.((.(((..((.((((((......(((((....)))))...))))))))...(((.((((((......)))))).)))))).)).))))....... ( -43.40) >DroSec_CAF1 131877 106 - 1 UCUUCAGCAGCACUCGACAGGUGGAGCUCUGCAAGGUGCACAUGCACCACUACCUGCCCGACGUGGCCUGCUUCGUCCAGCAGGACACUCGCGUCCUGCACUCAAC .....(((((((((((.(((((((.((...))..(((((....))))).)))))))..))).)))..))))).......(((((((......)))))))....... ( -44.50) >DroSim_CAF1 135119 106 - 1 UCUUCAGCAGCACUCGACAGGUGGAGCUGUGCAAGGUGCACAUGCACCACUACCUGCCCGACGUGGCCUGCUUUGUCCAGCAGGACACUCGCGUCCUGCACUCCAC .....(((((((((((.(((((((.((...))..(((((....))))).)))))))..))).)))..))))).......(((((((......)))))))....... ( -44.50) >DroEre_CAF1 136178 106 - 1 UCUUCAGCAGCACUCGACAGGAGGAGCUCUGCCAGGUGCACAUGCACCACUACCUGCCCGACGUGGCCUGCUUCACCCAGCAGGACACGCGCGUCCUGCACUCCAC .....(((((((((((.((((.((.......)).(((((....)))))....))))..))).)))..))))).......(((((((......)))))))....... ( -40.20) >DroYak_CAF1 135882 106 - 1 UCUUCAGCAGCACGCGGCAGGUGGAGCUCUGCCAGGUGCACAUGCACCACUACCUGCCCGACGUGGCCUGCUUCGCCCAGCAGGACACUCGCGUCCUGCACUCCAC ......((((.(((((((((((((.((...))..(((((....))))).))))))))).((.(((.((((((......)))))).))))))))).))))....... ( -53.50) >DroAna_CAF1 185944 106 - 1 UUAUCAACACCACUCGCCAGGUAGUGCUCAGCCAGGUGCACAUGCACCGCUAUCAUCCUGACGUCGCCUGUUUCACGCAGCGGGAGGCCCGUGUCCUGCACUCUAC ...............(((((((..((.((((...(((((....))))).........))))))..)))))......)).((((((.(....).))))))....... ( -30.50) >consensus UCUUCAGCAGCACUCGACAGGUGGAGCUCUGCAAGGUGCACAUGCACCACUACCUGCCCGACGUGGCCUGCUUCGCCCAGCAGGACACUCGCGUCCUGCACUCCAC .....(((((((((((.(((((((.((...))..(((((....))))).)))))))..))).)))..))))).......(((((((......)))))))....... (-36.15 = -36.82 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:09 2006