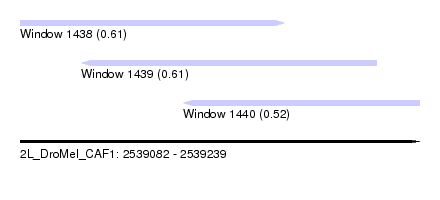
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,539,082 – 2,539,239 |
| Length | 157 |
| Max. P | 0.612021 |

| Location | 2,539,082 – 2,539,186 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -22.23 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2539082 104 + 22407834 AUCGAAUCGAACUGCA-AAUCUCAAUGCUGCAAUUCGUGCUCGGCAAGCAAACAUUGCAUAAAUUGCUUGGCAAUGGCCACUAAAAUGCCACAUGGAAUGGCAGC ................-.........(((((..((((((...((((..........(((.....))).((((....))))......)))).))))))...))))) ( -25.90) >DroSec_CAF1 118650 104 + 1 AUCGAAUCGAACUGCA-AAUCUCAAUGCUGCAAUUCGUGCUCGGCAAGCAAACAUUGCAUAAAUUGCUUGGCAAUGGCCACUAAAAUGCCACAUGGAAUGGCAGC ................-.........(((((..((((((...((((..........(((.....))).((((....))))......)))).))))))...))))) ( -25.90) >DroSim_CAF1 121733 104 + 1 AUCGAAUCGAACUGCA-AAUCUCAAUGCUGCAAUUCGUGCUCGGCAAGCAAACAUUGCAUAAAUUGCUUGGCAAUGGCCACUAAAAUGCCACAUGGAAUGGCAGC ................-.........(((((..((((((...((((..........(((.....))).((((....))))......)))).))))))...))))) ( -25.90) >DroEre_CAF1 121880 104 + 1 AUCGAAUCGAACUGCA-AAUCUCAAUGCUGCAAUUCGUGCUCGGCAAGCAAACAUUGCAUAAAUUGCUUGGCAAUGGCCACUAAAAUGCCACAUGGAAUGGCAUC ...((..((((.((((-...........)))).))))...))(((..(((.....)))....(((((...))))).)))......((((((.......)))))). ( -23.90) >DroYak_CAF1 122279 104 + 1 AUCGAAUCGAACUGCA-AAUCUCAAUGCUGCAAAUCGUGCUCGGCAAGCAAACAUUGCAUAAAUUGCUUGGCAAUGGCCACUAAAAUGCCACAUGGAAUGGCAUC ...((..(((..((((-...........))))..)))...))(((..(((.....)))....(((((...))))).)))......((((((.......)))))). ( -22.10) >DroAna_CAF1 154583 99 + 1 AUCGAAG------GCACAAUCUCACUGCUACAAUUCGUGCUCGGCAAGCAAACAUUGCAUAAAUUGCUUGGCAAUGGCCACUAAAAUGCCACAUGGAAUGGCAGC .......------...........((((((...((((((...((((..........(((.....))).((((....))))......)))).)))))).)))))). ( -24.30) >consensus AUCGAAUCGAACUGCA_AAUCUCAAUGCUGCAAUUCGUGCUCGGCAAGCAAACAUUGCAUAAAUUGCUUGGCAAUGGCCACUAAAAUGCCACAUGGAAUGGCAGC ..........................(((((..((((((...((((..........(((.....))).((((....))))......)))).))))))...))))) (-22.23 = -22.90 + 0.67)

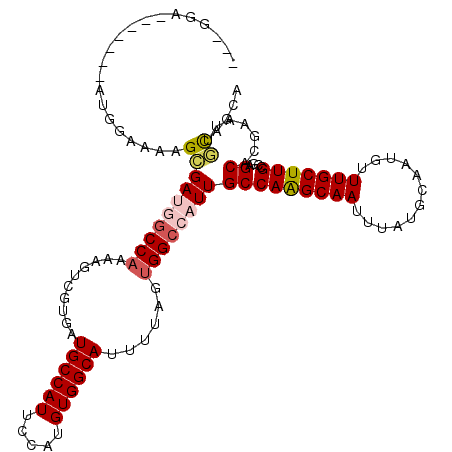
| Location | 2,539,106 – 2,539,222 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -23.23 |
| Energy contribution | -23.71 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2539106 116 - 22407834 ---GGAAAACUGGAUGGAAAAGCGAUGGCCAAAAGUCGUGCUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGCAAUUUAUGCAAUGUUUGCUUGCCGAGCACGAAUUGCAGCA ---..................((((((((((......((((..(........)..)))).....))))))))))...((((((..(((...((......))...)))..)))))).... ( -37.50) >DroPse_CAF1 151860 100 - 1 -------------------UGGGGAAAGCCAAGACUAGUGUUGCCAUUCCAUGUGGCAUUUUAUUGGCCAUUGCCAGGCAAUUUAUGCAAUGUUUGCUUGCCGAGCACGAACGCCAGCU -------------------(((.(...((((((((....)))(((((.....)))))......)))))...(((..(((((.....(((.....))))))))..)))....).)))... ( -29.50) >DroEre_CAF1 121904 119 - 1 UAGGGAAAACUGGAUGGAAAAGCGAUGGCCAAAAGUCGUGAUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGCAAUUUAUGCAAUGUUUGCUUGCCGAGCACGAAUUGCAGCA (((......))).........((((((((((........((((((((.....))))))))....))))))))))...((((((..(((...((......))...)))..)))))).... ( -40.40) >DroYak_CAF1 122303 119 - 1 UAGGGAAAACUGGAUGGAAAAGCGAUGGCCAAAAGUCGUGAUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGCAAUUUAUGCAAUGUUUGCUUGCCGAGCACGAUUUGCAGCA (((......)))..(((....((((((((((........((((((((.....))))))))....))))))))))(((((((............)))))))))).(((.....))).... ( -39.00) >DroAna_CAF1 154602 112 - 1 UAUGGA-------AUGGGUAAGAGAUGUCCCUGAGUCGUGCUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGCAAUUUAUGCAAUGUUUGCUUGCCGAGCACGAAUUGUAGCA ((((((-------(((((((...(((........))).)))..))))))))))..((.......((((....)))).((((((..(((...((......))...)))..)))))).)). ( -32.50) >DroPer_CAF1 156562 100 - 1 -------------------UGGGGAAAGCCAAGACUAGUGUUGCCAUUCCAUGUGGCAUUUUAUUGGCCAUUGCCAGGCAAUUUAUGCAAUGUUUGCUUGCCGAGCACGAACGCCAGCU -------------------(((.(...((((((((....)))(((((.....)))))......)))))...(((..(((((.....(((.....))))))))..)))....).)))... ( -29.50) >consensus ___GGA_______AUGGAAAAGCGAUGGCCAAAAGUCGUGAUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGCAAUUUAUGCAAUGUUUGCUUGCCGAGCACGAAUUGCAGCA .....................((((((((((..........((((((.....))))))......))))))))(((((((((............)))))))....)).......)).... (-23.23 = -23.71 + 0.47)
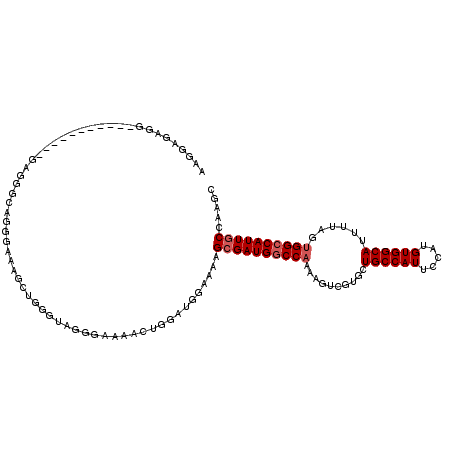
| Location | 2,539,146 – 2,539,239 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -21.61 |
| Energy contribution | -22.11 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2539146 93 - 22407834 AAGGAGAGG-----------GAGGGCAG--------------GGAAAACUGGAUGGAAAAGCGAUGGCCAAAAGUCGUGCUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGC .........-----------...(((((--------------((...((((((......(((.(((((.....))))))))(((((.....)))))..))))))..)).))))).... ( -28.30) >DroSec_CAF1 118714 107 - 1 AAGGAGAGG-----------GAGGGCAGGGAAAGCUGGGCAGGGAAAACUGGAUGGAAAAGCGAUGGCCAAAAGUCGUGCUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGC .........-----------.............(((.(((((((...((((((......(((.(((((.....))))))))(((((.....)))))..))))))..)).))))).))) ( -31.20) >DroSim_CAF1 121797 107 - 1 AAGGAGAGG-----------GAGGGCAGGGAAAGCUGGGCAGGGAAAACUGGAUGGAAAAGCGAUGGCCAAAAGUCGUGCUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGC .........-----------.............(((.(((((((...((((((......(((.(((((.....))))))))(((((.....)))))..))))))..)).))))).))) ( -31.20) >DroEre_CAF1 121944 107 - 1 AAGGAGAUG-----------GAGGGCGGGGAAAACUGGGUAGGGAAAACUGGAUGGAAAAGCGAUGGCCAAAAGUCGUGAUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGC .........-----------....((..(.....(((..(((......)))..)))....((((((((((........((((((((.....))))))))....)))))))))))..)) ( -31.80) >DroYak_CAF1 122343 118 - 1 AAGCAGUGGGGAGGGGAGGGAAGGGCAGGAAAAUCGGGGUAGGGAAAACUGGAUGGAAAAGCGAUGGCCAAAAGUCGUGAUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGC ..((.............................((.(..(((......)))..).))...((((((((((........((((((((.....))))))))....))))))))))...)) ( -32.20) >DroAna_CAF1 154642 100 - 1 GAGAAAACA-----------UAGAGCGUCUAAAUCGGGAUAUGGA-------AUGGGUAAGAGAUGUCCCUGAGUCGUGCUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGC .........-----------....((..(((((..(..(((((((-------(((((((...(((........))).)))..)))))))))))..)..)))))((((....)))).)) ( -26.10) >consensus AAGGAGAGG___________GAGGGCAGGGAAAGCUGGGUAGGGAAAACUGGAUGGAAAAGCGAUGGCCAAAAGUCGUGCUGCCAUUCCAUGUGGCAUUUUAGUGGCCAUUGCCAAGC ............................................................((((((((((..........((((((.....))))))......))))))))))..... (-21.61 = -22.11 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:04 2006