| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,515,417 – 2,515,539 |
| Length | 122 |
| Max. P | 0.790681 |

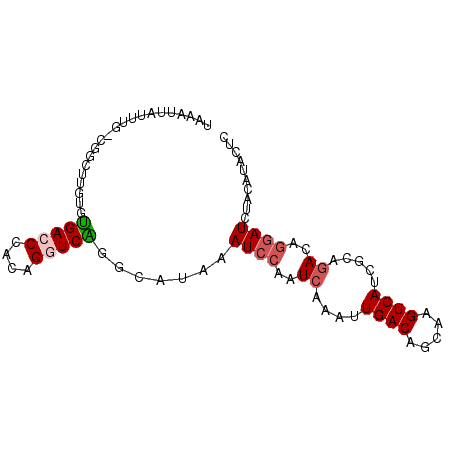
| Location | 2,515,417 – 2,515,507 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -19.91 |
| Consensus MFE | -9.44 |
| Energy contribution | -9.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2515417 90 + 22407834 UAAAUUAUUUGCCGGCUUGUGUGACCCACAGGUCAGGCAUAAAUCCAAUCAAAUUGACAGCAAGUCAACGCUGACAGGAUCUACAUACUC .........((((((((((((.....)))))))).))))...((((..(((..(((((.....)))))...)))..)))).......... ( -25.40) >DroPse_CAF1 118591 77 + 1 UAAAUUAU-------------UGACCCAUGGCUCAGGCAUAAAUCCAAUCAACUUGACAGCAAGUCAUCGUGGACAGGAUCUGCAUAUUA .......(-------------(((.(....).))))(((...((((..((.((.((((.....))))..)).))..)))).)))...... ( -13.60) >DroSec_CAF1 95200 90 + 1 UAAAUUAUUUGCCGGCUUGUGUGACCCACAGGUCGGGCAUAAAUCCAAUCAAAUUGACAGCAAGUCAAUGCUGACAGGAUCUACAUACUC .........((((((((((((.....)))))))).))))...((((..(((.((((((.....))))))..)))..)))).......... ( -28.10) >DroEre_CAF1 98562 90 + 1 UAAAUUAUGUGUUGGCUUGUGUGACCCACAGGUCAGGCAUAAAUCCAAUCAAAUUGACAGCAAGUCACGGCAGACAGGAUCUACAUACUC ....((((((.((((((((((.....))))))))))))))))((((..((....((((.....)))).....))..)))).......... ( -22.70) >DroAna_CAF1 117039 90 + 1 UACAUUAUUUUUCCAAAUGAGGGACCCACGGGUCUCGCAUAAAUCCAAUCAAAUUGACAGCAAGUCAUCAUAAAUGACAUUUCCAUAUUC ..((((..........(((.((((((....)))))).)))..............((((.....)))).....)))).............. ( -17.10) >DroPer_CAF1 117087 77 + 1 UAAAUUAU-------------UGACCCAUGGCUCAGGCAUAAAUCCAAUCAACCUGACAGCAAGUCAUCGUGGACAGGAUCUCCAUAUUA ........-------------((((.....(((((((...............)))))..))..))))..(((((.......))))).... ( -12.56) >consensus UAAAUUAUUUG_CGGCUUGUGUGACCCACAGGUCAGGCAUAAAUCCAAUCAAAUUGACAGCAAGUCAUCGCAGACAGGAUCUACAUACUC .....................(((((....))))).......((((..((....((((.....)))).....))..)))).......... ( -9.44 = -9.88 + 0.44)

| Location | 2,515,417 – 2,515,507 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.02 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
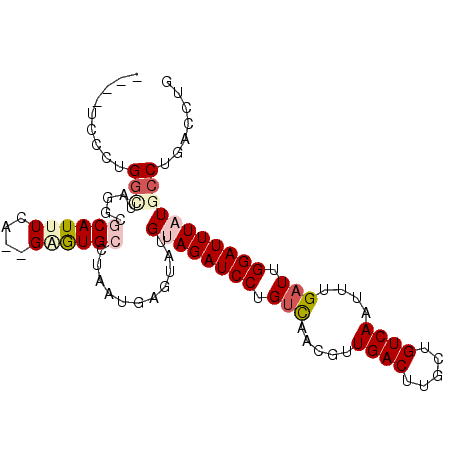
>2L_DroMel_CAF1 2515417 90 - 22407834 GAGUAUGUAGAUCCUGUCAGCGUUGACUUGCUGUCAAUUUGAUUGGAUUUAUGCCUGACCUGUGGGUCACACAAGCCGGCAAAUAAUUUA ......((((((((.(((((.((((((.....))))))))))).))))))))(((.(.(.((((.....)))).)).))).......... ( -29.00) >DroPse_CAF1 118591 77 - 1 UAAUAUGCAGAUCCUGUCCACGAUGACUUGCUGUCAAGUUGAUUGGAUUUAUGCCUGAGCCAUGGGUCA-------------AUAAUUUA ......((((((((.(((.((..((((.....)))).)).))).)))))..))).(((.(....).)))-------------........ ( -18.90) >DroSec_CAF1 95200 90 - 1 GAGUAUGUAGAUCCUGUCAGCAUUGACUUGCUGUCAAUUUGAUUGGAUUUAUGCCCGACCUGUGGGUCACACAAGCCGGCAAAUAAUUUA ......((((((((.(((((.((((((.....))))))))))).))))))))(((.(.(.((((.....)))).)).))).......... ( -29.50) >DroEre_CAF1 98562 90 - 1 GAGUAUGUAGAUCCUGUCUGCCGUGACUUGCUGUCAAUUUGAUUGGAUUUAUGCCUGACCUGUGGGUCACACAAGCCAACACAUAAUUUA ...(((((((((((.(((.....((((.....))))....))).)))))).((..(((((....)))))..)).......)))))..... ( -23.50) >DroAna_CAF1 117039 90 - 1 GAAUAUGGAAAUGUCAUUUAUGAUGACUUGCUGUCAAUUUGAUUGGAUUUAUGCGAGACCCGUGGGUCCCUCAUUUGGAAAAAUAAUGUA ..((((.......(((...((((((((.....))))........(((((((((.(....)))))))))).)))).))).......)))). ( -15.84) >DroPer_CAF1 117087 77 - 1 UAAUAUGGAGAUCCUGUCCACGAUGACUUGCUGUCAGGUUGAUUGGAUUUAUGCCUGAGCCAUGGGUCA-------------AUAAUUUA ......((.(((((.(((.((..((((.....)))).)).))).)))))....))(((.(....).)))-------------........ ( -18.20) >consensus GAAUAUGUAGAUCCUGUCAACGAUGACUUGCUGUCAAUUUGAUUGGAUUUAUGCCUGACCCGUGGGUCACACAAGCCG_CAAAUAAUUUA ........((((((.(((.....((((.....))))....))).))))))......((((....))))...................... (-15.07 = -15.02 + -0.05)

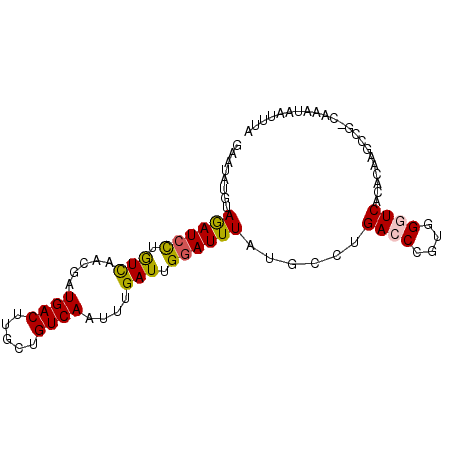
| Location | 2,515,445 – 2,515,539 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.90 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2515445 94 - 22407834 ----UCCCUGACUAGGCGCAUUUCA--GAGUGCCUAAUGAGUAUGUAGAUCCUGUCAGCGUUGACUUGCUGUCAAUUUGAUUGGAUUUAUGCCUGACCUG ----...((.(.(((((((......--..))))))).).))...((((((((.(((((.((((((.....))))))))))).)))))))).......... ( -30.90) >DroPse_CAF1 118606 100 - 1 AUGAUUCAUGGGGAAAUCCAUAUCCGGGAAUGCCUAAUUAAUAUGCAGAUCCUGUCCACGAUGACUUGCUGUCAAGUUGAUUGGAUUUAUGCCUGAGCCA .((..(((.((.((((((((....(((((.(((.((.....)).)))..)))))((.((..((((.....)))).)).)).))))))).).)))))..)) ( -25.60) >DroSec_CAF1 95228 94 - 1 ----UCCCUGGUUAGGCGCAUUUCA--GAGUGCCUAAUGAGUAUGUAGAUCCUGUCAGCAUUGACUUGCUGUCAAUUUGAUUGGAUUUAUGCCCGACCUG ----...((.(((((((((......--..))))))))).))...((((((((.(((((.((((((.....))))))))))).)))))))).......... ( -35.40) >DroEre_CAF1 98590 94 - 1 ----UCCCUGGCUAGGCGCAUUUCA--GAGUGCCUAAUGAGUAUGUAGAUCCUGUCUGCCGUGACUUGCUGUCAAUUUGAUUGGAUUUAUGCCUGACCUG ----.....((((((((((......--..)))))))........((((((((.(((.....((((.....))))....))).)))))))))))....... ( -30.60) >DroYak_CAF1 98642 94 - 1 ----UCCCUGGUUACGCGCAUUUCA--GGGUGCCUAAUGAGCAUGUAGAUCCUGCUAAAGUUGACUUGACGUCAAUUUGAUUGGAUUUAUGCCUGACCUG ----.....(((((.(((((((...--.)))))...........((((((((.....((((((((.....))))))))....)))))))))).))))).. ( -28.20) >DroPer_CAF1 117102 100 - 1 AUGAUUCAUGGGUAAAUCCAUAUCCGGGAAUGCCUAAUUAAUAUGGAGAUCCUGUCCACGAUGACUUGCUGUCAGGUUGAUUGGAUUUAUGCCUGAGCCA .((..(((.(.(((((((((....(((((...((..........))...)))))((.((..((((.....)))).)).)).))))))))).).)))..)) ( -27.60) >consensus ____UCCCUGGCUAGGCGCAUUUCA__GAGUGCCUAAUGAGUAUGUAGAUCCUGUCAACGUUGACUUGCUGUCAAUUUGAUUGGAUUUAUGCCUGACCUG .........(((.....((((((....))))))...........((((((((.(((.....((((.....))))....))).)))))))))))....... (-14.79 = -15.90 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:53 2006