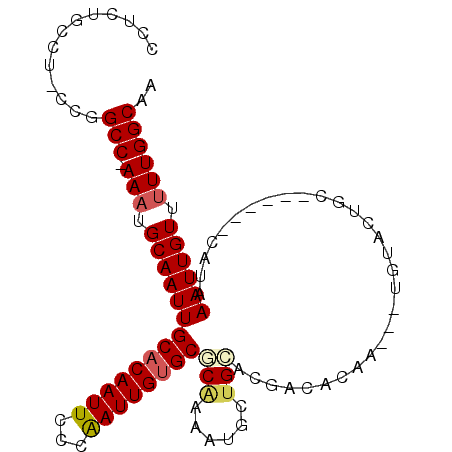
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
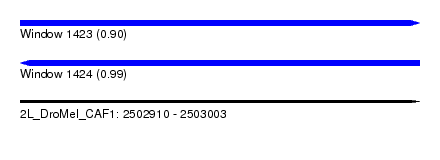
| Location | 2,502,910 – 2,503,003 |
| Length | 93 |
| Max. P | 0.991188 |

| Location | 2,502,910 – 2,503,003 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.03 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -14.95 |
| Energy contribution | -14.87 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2502910 93 + 22407834 UUGCCAAAAACAAUUUAUG------GCAGCACA---CUGUGUCGUUCAGAAUUUUGCGCACAAUUGAGAAUUGUGCAAUUGCAUUUUGGCCGG-CGGAAGACG ((((((.((.....)).))------))))....---((.(((((..(((((...((((((((((.....)))))))....))))))))..)))-))..))... ( -25.20) >DroPse_CAF1 96227 103 + 1 UUGCCAAAAACAAUUUAUGCUUCUCGCAGUACAAUAUUGUGUCGUGCAACAUUUUGCGCGCAAUCGAGAAUUGUGCAAUUGCAUUUGGGCCAAAAAGCAGAGG ...(((((..(((((..(((.....)))(((((((.(((((.(((((((....)))))))))..)))..))))))))))))..)))))((......))..... ( -27.30) >DroYak_CAF1 85263 93 + 1 UUGCCAAAAACAAUUUAUG------GCAGCACA---CUGUGUCGUUCUGAAUUUUGCGCACAAUUGGGAAUUGUGCAAUUGCAUUUUGGCCGG-CGCAAGACG ((((((.((.....)).))------))))....---.(((((((..(..((...((((((((((.....)))))))....))).))..).)))-))))..... ( -26.10) >DroMoj_CAF1 126203 90 + 1 UUGCCAA-AACAAUUUAUG------UUCGCA-----UUUUGGCCAACAAUAUUUCGUGCACAAUUGGAAAUUGUGCAAUUGCAUUU-GGCCAGCAGGCAGAGC (((((..-((((.....))------)).((.-----...(((((((..........((((((((.....)))))))).......))-))))))).)))))... ( -26.93) >DroAna_CAF1 105236 87 + 1 CUGCCAAAAACAAUUUAUG------CAAGUA-----UUGUGUAUUGCAGCGUUUUGCGCACAAUUGGGAAUUGUGCAAUUGCAUUU-GGCUGC-AG---GCUG ..(((..........((((------((....-----.)))))).((((((....((((((((((.....)))))))....)))...-.)))))-))---)).. ( -26.90) >DroPer_CAF1 93592 103 + 1 UUGCCAAAAACAAUUUAUGAUUCUCGCAGUACAAUAUUGUGUCGUGCAACAUUUUGCGCACAAUCGAGAAUUGUGCAAUUGCAUUUGGGCCAAAAAGCAGAGG ...(((((..(((((((..((((((((((((...))))))((.((((((....))))))))....))))))..)).)))))..)))))((......))..... ( -29.10) >consensus UUGCCAAAAACAAUUUAUG______GCAGCACA___UUGUGUCGUGCAACAUUUUGCGCACAAUUGAGAAUUGUGCAAUUGCAUUU_GGCCAG_AGGCAGAGG ..((((((..(((((.............(((.....((((.....)))).....)))(((((((.....))))))))))))..))).)))............. (-14.95 = -14.87 + -0.08)

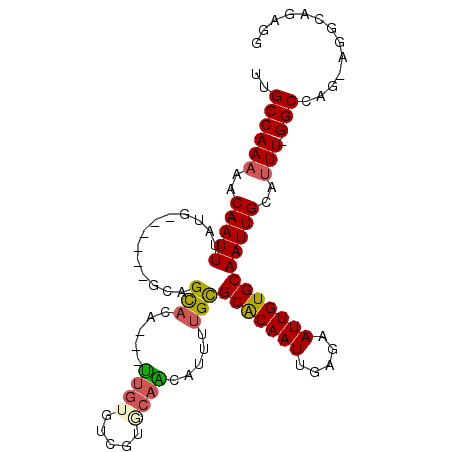
| Location | 2,502,910 – 2,503,003 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.03 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.13 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2502910 93 - 22407834 CGUCUUCCG-CCGGCCAAAAUGCAAUUGCACAAUUCUCAAUUGUGCGCAAAAUUCUGAACGACACAG---UGUGCUGC------CAUAAAUUGUUUUUGGCAA .........-...(((((((.(((((((((((((.....)))))))(((..((.(((.......)))---.))..)))------....))))))))))))).. ( -26.30) >DroPse_CAF1 96227 103 - 1 CCUCUGCUUUUUGGCCCAAAUGCAAUUGCACAAUUCUCGAUUGCGCGCAAAAUGUUGCACGACACAAUAUUGUACUGCGAGAAGCAUAAAUUGUUUUUGGCAA .....(((....)))(((((.((((((......((((((....((.((((....)))).))(((......)))....)))))).....)))))).)))))... ( -24.70) >DroYak_CAF1 85263 93 - 1 CGUCUUGCG-CCGGCCAAAAUGCAAUUGCACAAUUCCCAAUUGUGCGCAAAAUUCAGAACGACACAG---UGUGCUGC------CAUAAAUUGUUUUUGGCAA .........-...(((((((.(((((((((((((.....)))))))........(((.((......)---)...))).------....))))))))))))).. ( -24.10) >DroMoj_CAF1 126203 90 - 1 GCUCUGCCUGCUGGCC-AAAUGCAAUUGCACAAUUUCCAAUUGUGCACGAAAUAUUGUUGGCCAAAA-----UGCGAA------CAUAAAUUGUU-UUGGCAA ....((((.(((((((-(...(((((((((((((.....))))))))......)))))))))))...-----.))(((------((.....))))-).)))). ( -28.50) >DroAna_CAF1 105236 87 - 1 CAGC---CU-GCAGCC-AAAUGCAAUUGCACAAUUCCCAAUUGUGCGCAAAACGCUGCAAUACACAA-----UACUUG------CAUAAAUUGUUUUUGGCAG ....---..-...(((-(((.(((((((((((((.....)))))))((.....))(((((.......-----...)))------))..)))))).)))))).. ( -25.30) >DroPer_CAF1 93592 103 - 1 CCUCUGCUUUUUGGCCCAAAUGCAAUUGCACAAUUCUCGAUUGUGCGCAAAAUGUUGCACGACACAAUAUUGUACUGCGAGAAUCAUAAAUUGUUUUUGGCAA .....(((....)))(((((.((((((.....(((((((...(((((....((((((.......)))))))))))..)))))))....)))))).)))))... ( -27.00) >consensus CCUCUGCCU_CCGGCC_AAAUGCAAUUGCACAAUUCCCAAUUGUGCGCAAAAUGCUGCACGACACAA___UGUACUGC______CAUAAAUUGUUUUUGGCAA .............(((.(((.((((((((((((((...))))))))(((......)))..............................)))))).)))))).. (-16.80 = -17.13 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:48 2006