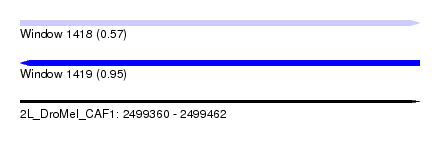
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,499,360 – 2,499,462 |
| Length | 102 |
| Max. P | 0.954989 |

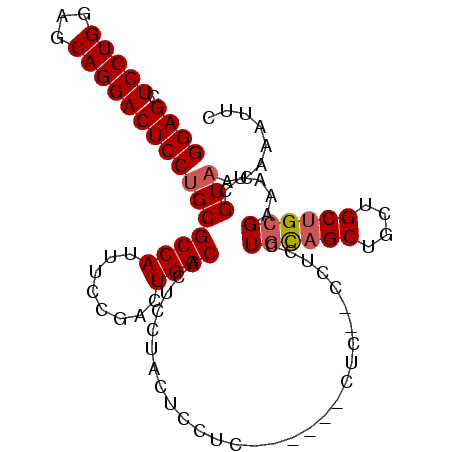
| Location | 2,499,360 – 2,499,462 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 86.03 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -24.10 |
| Energy contribution | -25.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2499360 102 + 22407834 GAAUUUGUUACGAUUUCGCAGCAGCAGCUACAGGAGAAGGAGGAACAGGAGGAGGAGGAAUGCCAGUCGGAAAUGGCGCAGGAGUCCUGCUCCAGGAGCUCC ...((((((.(..((((..(((....)))......))))..).)))))).((((.((((...((.((((....))))...))..)))).))))......... ( -27.50) >DroSec_CAF1 78291 90 + 1 GAAUUUGUUACGAUUUCGCAGCAGCAGCUGCAGGAGG------------AGGAGUAGGAGUGCCAGUCGGAAAUGGCGCAGGAGUCCUGCUCCAGGAGCUCC (((.(((...))).)))(((((....))))).((((.------------.(((((((((((((((........)))))).....))))))))).....)))) ( -36.80) >DroSim_CAF1 81674 96 + 1 GAAUUUGUUACGAUUUCGCAGCAGCAGCUGCAGGAGGAGGAG------GAGGAGUAGGAGUGCCAGUCGGAAAUGGCGCAGGAGUCCUGCUCCAGGAGCUCC (((.(((...))).)))(((((....)))))....((((...------..(((((((((((((((........)))))).....))))))))).....)))) ( -38.00) >DroYak_CAF1 81728 94 + 1 GAAUUUGUUACGAUUUCGCAGCAGCAGCAGGAGGAG---UGG-----UGAAGUGGAGCAGUGCCAGUCGGAAAUGGCGCUGGAGUCCUGCGCCAGGACCUCC ....(((((.((....)).))))).....(((((..---(((-----((....(((.((((((((........))))))))...)))..)))))...))))) ( -34.90) >consensus GAAUUUGUUACGAUUUCGCAGCAGCAGCUGCAGGAGG__GAG______GAGGAGGAGGAGUGCCAGUCGGAAAUGGCGCAGGAGUCCUGCUCCAGGAGCUCC (((.(((...))).)))(((((....)))))............................((((((........)))))).(((((((((...))))).)))) (-24.10 = -25.10 + 1.00)

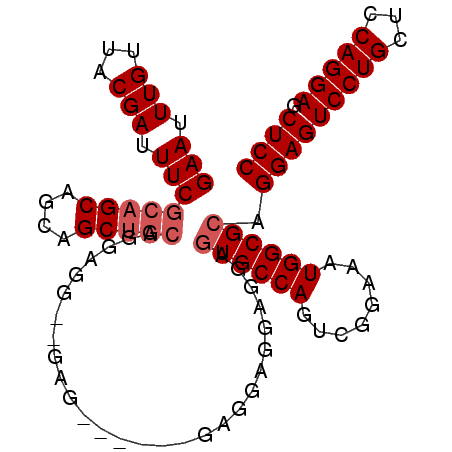
| Location | 2,499,360 – 2,499,462 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 86.03 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2499360 102 - 22407834 GGAGCUCCUGGAGCAGGACUCCUGCGCCAUUUCCGACUGGCAUUCCUCCUCCUCCUGUUCCUCCUUCUCCUGUAGCUGCUGCUGCGAAAUCGUAACAAAUUC ((((.....((((((((((....).((((........))))...........))))))))).....))))((((((....))))))................ ( -31.40) >DroSec_CAF1 78291 90 - 1 GGAGCUCCUGGAGCAGGACUCCUGCGCCAUUUCCGACUGGCACUCCUACUCCU------------CCUCCUGCAGCUGCUGCUGCGAAAUCGUAACAAAUUC ((((.....((((((((...)))))((((........))))..))).....))------------))...((((((....))))))................ ( -30.50) >DroSim_CAF1 81674 96 - 1 GGAGCUCCUGGAGCAGGACUCCUGCGCCAUUUCCGACUGGCACUCCUACUCCUC------CUCCUCCUCCUGCAGCUGCUGCUGCGAAAUCGUAACAAAUUC ((((.....((((((((...)))))((((........))))..))).....)))------).........((((((....))))))................ ( -30.50) >DroYak_CAF1 81728 94 - 1 GGAGGUCCUGGCGCAGGACUCCAGCGCCAUUUCCGACUGGCACUGCUCCACUUCA-----CCA---CUCCUCCUGCUGCUGCUGCGAAAUCGUAACAAAUUC (((((...(((.(.((.....(((.((((........)))).))).....)).).-----)))---..))))).((....))((((....))))........ ( -27.30) >consensus GGAGCUCCUGGAGCAGGACUCCUGCGCCAUUUCCGACUGGCACUCCUACUCCUC______CUC__CCUCCUGCAGCUGCUGCUGCGAAAUCGUAACAAAUUC ((((.(((((...)))))))))(((((((........)))).............................((((((....)))))).....)))........ (-23.94 = -24.50 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:44 2006