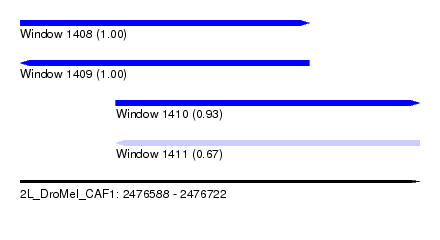
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,476,588 – 2,476,722 |
| Length | 134 |
| Max. P | 0.999938 |

| Location | 2,476,588 – 2,476,685 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -36.86 |
| Consensus MFE | -35.40 |
| Energy contribution | -35.08 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.65 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.68 |
| SVM RNA-class probability | 0.999938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2476588 97 + 22407834 UUAUCUGUUUGGCUCGUUGCCGUUUUUUGUAUAAUUU----UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCU ..........(((.....)))................----...((((((((..((((..((((..((((........))))..)))))))).)))))))) ( -36.50) >DroSec_CAF1 55851 97 + 1 UUAUCUUUUUGGCUCGUUGCCGUUUUUUGUAUAAUUU----UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCU ..........(((.....)))................----...((((((((..((((..((((..((((........))))..)))))))).)))))))) ( -36.50) >DroSim_CAF1 58853 97 + 1 UUAUCUGUUUGGCUCGUUGCCGUUUUUUGUAUAAUUU----UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCU ..........(((.....)))................----...((((((((..((((..((((..((((........))))..)))))))).)))))))) ( -36.50) >DroEre_CAF1 58054 96 + 1 UUAUCUGUUUGGCUUGUUGCCGUU-UUUGUAUAAUUU----UUUAGGUGCUGCUUGCUGCUGGGUGUGACAAAUUAACGUCGUACCCAAGCGACAGCACCU .(((..(..((((.....))))..-)..)))......----...((((((((.(((((..((((((((((........))))))))))))))))))))))) ( -37.60) >DroYak_CAF1 58342 101 + 1 UUAUCUGUUUGGCUUGUUGCCGUUUUUUGUAUAAUUUUUUUUUUGGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCAACAGCACCU ..........(((.....))).......................((((((((..((((..((((..((((........))))..)))))))).)))))))) ( -37.20) >consensus UUAUCUGUUUGGCUCGUUGCCGUUUUUUGUAUAAUUU____UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCU ..........(((.....))).......................((((((((..((((..((((..((((........))))..)))))))).)))))))) (-35.40 = -35.08 + -0.32)

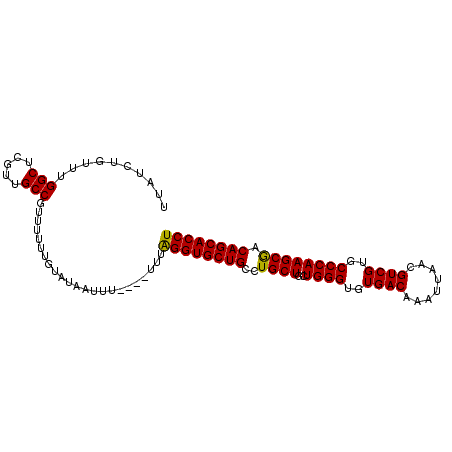
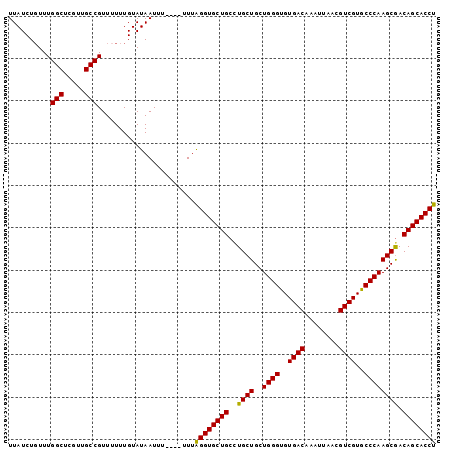
| Location | 2,476,588 – 2,476,685 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -32.78 |
| Energy contribution | -33.06 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -5.44 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2476588 97 - 22407834 AGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA----AAAUUAUACAAAAAACGGCAACGAGCCAAACAGAUAA (((((((((((((((((...((((......))))...))))..))).))))))))))...----................(((.....))).......... ( -34.80) >DroSec_CAF1 55851 97 - 1 AGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA----AAAUUAUACAAAAAACGGCAACGAGCCAAAAAGAUAA (((((((((((((((((...((((......))))...))))..))).))))))))))...----................(((.....))).......... ( -34.80) >DroSim_CAF1 58853 97 - 1 AGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA----AAAUUAUACAAAAAACGGCAACGAGCCAAACAGAUAA (((((((((((((((((...((((......))))...))))..))).))))))))))...----................(((.....))).......... ( -34.80) >DroEre_CAF1 58054 96 - 1 AGGUGCUGUCGCUUGGGUACGACGUUAAUUUGUCACACCCAGCAGCAAGCAGCACCUAAA----AAAUUAUACAAA-AACGGCAACAAGCCAAACAGAUAA (((((((((.((((((((..((((......))))..)))))..)))..)))))))))...----............-...(((.....))).......... ( -33.40) >DroYak_CAF1 58342 101 - 1 AGGUGCUGUUGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCCAAAAAAAAAAUUAUACAAAAAACGGCAACAAGCCAAACAGAUAA .((((((((((((((((...((((......))))...))))..)))).))))))))........................(((.....))).......... ( -32.30) >consensus AGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA____AAAUUAUACAAAAAACGGCAACGAGCCAAACAGAUAA (((((((((((((((((...((((......))))...))))..))).)))))))))).......................(((.....))).......... (-32.78 = -33.06 + 0.28)

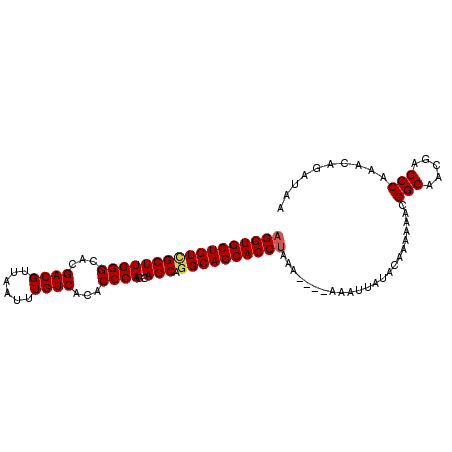
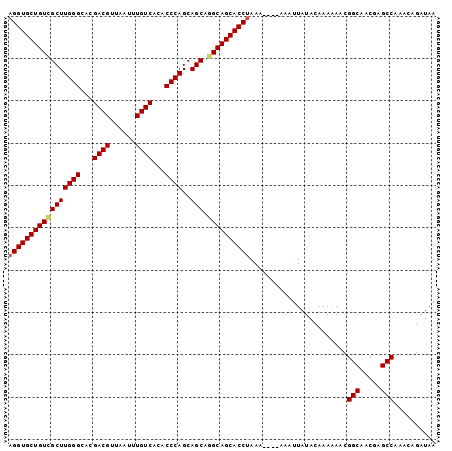
| Location | 2,476,620 – 2,476,722 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.64 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.35 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2476620 102 + 22407834 AAUUU----UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA--AAAUCAACAA-AAUCCUCAACGAAAUCGCGCGAC .((((----(((((((((((..((((..((((..((((........))))..)))))))).)))))))).)))--))))......-....................... ( -35.10) >DroSec_CAF1 55883 102 + 1 AAUUU----UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA--AAGUCAACAA-AAUCCUCAACGAAAUCGCGCGAC .((((----(((((((((((..((((..((((..((((........))))..)))))))).)))))))).)))--))))......-....................... ( -35.10) >DroSim_CAF1 58885 102 + 1 AAUUU----UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA--AAGUCAACAA-AAUCCUCAACGAAAUCGCGCGAC .((((----(((((((((((..((((..((((..((((........))))..)))))))).)))))))).)))--))))......-....................... ( -35.10) >DroEre_CAF1 58085 102 + 1 AAUUU----UUUAGGUGCUGCUUGCUGCUGGGUGUGACAAAUUAACGUCGUACCCAAGCGACAGCACCUCAAA--AAAUCAACAA-AAUCCUCAACGAAAUCGCGCGAC .((((----(((((((((((.(((((..((((((((((........))))))))))))))))))))))).)))--))))......-....................... ( -35.60) >DroYak_CAF1 58374 108 + 1 AAUUUUUUUUUUGGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCAACAGCACCUCACAGCAAAUCAACAAAAAUCCUCAGCGAAAUCGCGCGA- ............((((((((..((((..((((..((((........))))..)))))))).)))))))).........................(((....)))....- ( -36.70) >DroAna_CAF1 66057 77 + 1 AUUUU----UUU--GUGG----UGCUGCUGGGUGUGACAAAUUAACAUCGCGCCCAACUGCCAGCAU------------CAA----------CAACAAAAGCCCGCGAC .((((----.((--((((----(((((((((((((((..........)))))))))...).))))))------------).)----------))).))))......... ( -26.10) >consensus AAUUU____UUUAGGUGCUGCCUGCUGCUGGGUGUGACAAAUUAACGUCGUGCCCAAGCGACAGCACCUCAGA__AAAUCAACAA_AAUCCUCAACGAAAUCGCGCGAC ............((((((((..((((..((((((((((........)))))))))))))).))))))))........................................ (-25.24 = -25.35 + 0.11)


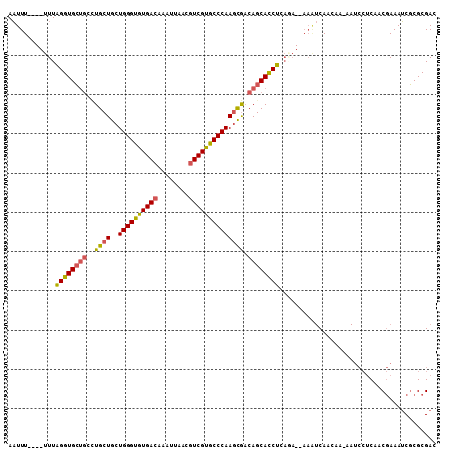
| Location | 2,476,620 – 2,476,722 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.64 |
| Mean single sequence MFE | -33.89 |
| Consensus MFE | -23.47 |
| Energy contribution | -25.05 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2476620 102 - 22407834 GUCGCGCGAUUUCGUUGAGGAUU-UUGUUGAUUU--UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA----AAAUU .....(((....))).(((((((-.....)))))--))..(((((((((((((((((...((((......))))...))))..))).))))))))))...----..... ( -35.50) >DroSec_CAF1 55883 102 - 1 GUCGCGCGAUUUCGUUGAGGAUU-UUGUUGACUU--UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA----AAAUU ((((.((((..((......))..-))))))))..--....(((((((((((((((((...((((......))))...))))..))).))))))))))...----..... ( -37.70) >DroSim_CAF1 58885 102 - 1 GUCGCGCGAUUUCGUUGAGGAUU-UUGUUGACUU--UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA----AAAUU ((((.((((..((......))..-))))))))..--....(((((((((((((((((...((((......))))...))))..))).))))))))))...----..... ( -37.70) >DroEre_CAF1 58085 102 - 1 GUCGCGCGAUUUCGUUGAGGAUU-UUGUUGAUUU--UUUGAGGUGCUGUCGCUUGGGUACGACGUUAAUUUGUCACACCCAGCAGCAAGCAGCACCUAAA----AAAUU .....((((..((......))..-)))).(((((--(((.(((((((((.((((((((..((((......))))..)))))..)))..))))))))))))----))))) ( -35.80) >DroYak_CAF1 58374 108 - 1 -UCGCGCGAUUUCGCUGAGGAUUUUUGUUGAUUUGCUGUGAGGUGCUGUUGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCCAAAAAAAAAAUU -(((((((....))).(..((((......))))..).))))((((((((((((((((...((((......))))...))))..)))).))))))))............. ( -35.80) >DroAna_CAF1 66057 77 - 1 GUCGCGGGCUUUUGUUG----------UUG------------AUGCUGGCAGUUGGGCGCGAUGUUAAUUUGUCACACCCAGCAGCA----CCAC--AAA----AAAAU ...((.(((........----------...------------..))).)).((((((.(.((((......)))).).))))))....----....--...----..... ( -20.82) >consensus GUCGCGCGAUUUCGUUGAGGAUU_UUGUUGAUUU__UCUGAGGUGCUGUCGCUUGGGCACGACGUUAAUUUGUCACACCCAGCAGCAGGCAGCACCUAAA____AAAUU .(((.(((....))))))......................(((((((((((((((((...((((......))))...))))..))).))))))))))............ (-23.47 = -25.05 + 1.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:36 2006