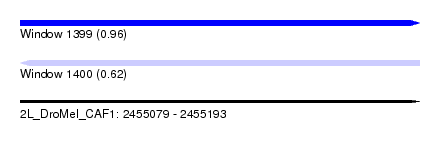
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,455,079 – 2,455,193 |
| Length | 114 |
| Max. P | 0.961512 |

| Location | 2,455,079 – 2,455,193 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -30.09 |
| Energy contribution | -30.70 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
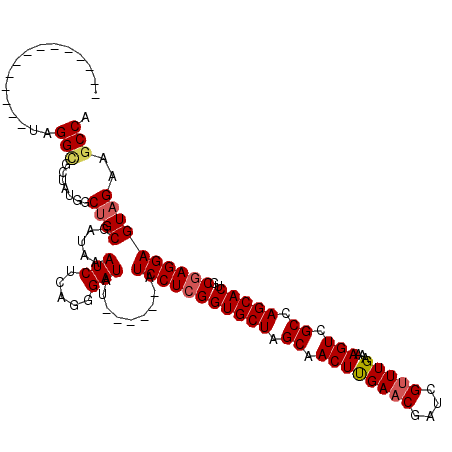
>2L_DroMel_CAF1 2455079 114 + 22407834 UGGCUUCUACUCCUCGCAGUGCUGGCGACUUUUCAAACGAUUGUUCGAGUUGCUAGCACCGAGGAU------AUAUCCCAGAGAUUCAUCGCCGCCAUAGCGCCCGUUGCCGCUCAUAUU (((..(.((.((((((..(((((((((((((..(((....)))...))))))))))))))))))))------).)..)))(((...((.((.(((....)))..)).))...)))..... ( -39.30) >DroGri_CAF1 50060 100 + 1 UGGCUUCUACUCCUCGCAGUGCUGGCGACUUUUCAAACGAUCGUUCAAGUUGCUAGCACCGAGGAU------AUAUCAACGAGAUUUAUCGCAGCUAUAGCACCAA-------------- (((((.....((((((..(((((((((((((....(((....))).))))))))))))))))))).------.......(((......))).))))).........-------------- ( -34.40) >DroWil_CAF1 51190 98 + 1 UGGCUUCUACUCCUGGCAGUGCUGGCGACUUUUCAAACGAUCGUUCAAGUUGCUAGCACCGAGGAU------AUAUCAGUUAGAUUUCUAGCAGCAAUAACGCC---------------- .(((......((((.(..(((((((((((((....(((....))).)))))))))))))).)))).------......(((((....))))).........)))---------------- ( -33.70) >DroMoj_CAF1 52334 100 + 1 UGGCUUCUACUCCUCGCAGUGCUGGCGACUUUUCAAACGAUCGUUCAAGUUGCUAGCACCGAGGAU------AUAUCGACCAGAUUUAUCGCAGCGAUAGCGCCCA-------------- .(((......((((((..(((((((((((((....(((....))).))))))))))))))))))).------.(((((....((....))....)))))..)))..-------------- ( -38.20) >DroAna_CAF1 42821 111 + 1 UGGCUUCUACUCCUCGCAGUGCUGGCGACUUUUCAAACGAUCGUUCGAGUUGCUAGCACCGAGGAC------AUAUCCCUGAGAUUCAUCGCAGCCAUAGCGCCU---CCUCCUAAGAUA .(((......((((((..(((((((((((((....(((....))).))))))))))))))))))).------.(((..(((.((....)).)))..)))..))).---............ ( -36.60) >DroPer_CAF1 37388 116 + 1 UGGAUUCUACUCCUCGCAGUGCUGGCGACUUCUCAACCGAUCGUUCAAGUUGCUAGCACCGAGGAUACGAGUAUAUCCCAGAGAUUCAUCGCAGCCAUAGCGCCUA--CCCGCACUGA-- .((((.((..((((((..(((((((((((((...(((.....))).)))))))))))))))))))....))...))))(((.((....)).........(((....--..))).))).-- ( -37.30) >consensus UGGCUUCUACUCCUCGCAGUGCUGGCGACUUUUCAAACGAUCGUUCAAGUUGCUAGCACCGAGGAU______AUAUCCCAGAGAUUCAUCGCAGCCAUAGCGCCCA______________ .(((......((((((..(((((((((((((....(((....))).)))))))))))))))))))............................((....)))))................ (-30.09 = -30.70 + 0.61)

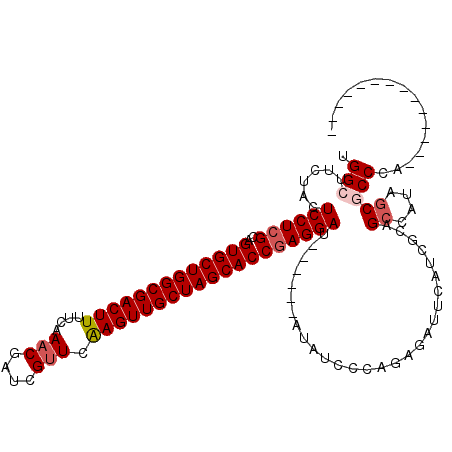
| Location | 2,455,079 – 2,455,193 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -28.26 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2455079 114 - 22407834 AAUAUGAGCGGCAACGGGCGCUAUGGCGGCGAUGAAUCUCUGGGAUAU------AUCCUCGGUGCUAGCAACUCGAACAAUCGUUUGAAAAGUCGCCAGCACUGCGAGGAGUAGAAGCCA ......((((.(....).)))).((((.((.....(((.....)))..------.(((((((((((.((.((((((((....))))))...)).)).)))))..))))))))....)))) ( -37.20) >DroGri_CAF1 50060 100 - 1 --------------UUGGUGCUAUAGCUGCGAUAAAUCUCGUUGAUAU------AUCCUCGGUGCUAGCAACUUGAACGAUCGUUUGAAAAGUCGCCAGCACUGCGAGGAGUAGAAGCCA --------------.((((.((((..(.((((......)))).)....------.(((((((((((.((.((((((((....))))...)))).)).)))))..))))))))))..)))) ( -31.40) >DroWil_CAF1 51190 98 - 1 ----------------GGCGUUAUUGCUGCUAGAAAUCUAACUGAUAU------AUCCUCGGUGCUAGCAACUUGAACGAUCGUUUGAAAAGUCGCCAGCACUGCCAGGAGUAGAAGCCA ----------------(((.......((((.....(((.....)))..------.(((((((((((.((.((((((((....))))...)))).)).)))))))..))))))))..))). ( -29.90) >DroMoj_CAF1 52334 100 - 1 --------------UGGGCGCUAUCGCUGCGAUAAAUCUGGUCGAUAU------AUCCUCGGUGCUAGCAACUUGAACGAUCGUUUGAAAAGUCGCCAGCACUGCGAGGAGUAGAAGCCA --------------..(((..((((((((.((....))))).))))).------.(((((((((((.((.((((((((....))))...)))).)).)))))..))))))......))). ( -33.10) >DroAna_CAF1 42821 111 - 1 UAUCUUAGGAGG---AGGCGCUAUGGCUGCGAUGAAUCUCAGGGAUAU------GUCCUCGGUGCUAGCAACUCGAACGAUCGUUUGAAAAGUCGCCAGCACUGCGAGGAGUAGAAGCCA ..(((.....))---)(((.((((..(((.((....)).)))......------.(((((((((((.((.((((((((....))))))...)).)).)))))..))))))))))..))). ( -34.90) >DroPer_CAF1 37388 116 - 1 --UCAGUGCGGG--UAGGCGCUAUGGCUGCGAUGAAUCUCUGGGAUAUACUCGUAUCCUCGGUGCUAGCAACUUGAACGAUCGGUUGAGAAGUCGCCAGCACUGCGAGGAGUAGAAUCCA --.((((((.((--(.((((((..((.(((((((.(((.....))).)).))))).))..))))))..(((((.((....))))))).......))).))))))...(((......))). ( -37.90) >consensus ______________UAGGCGCUAUGGCUGCGAUAAAUCUCAGGGAUAU______AUCCUCGGUGCUAGCAACUUGAACGAUCGUUUGAAAAGUCGCCAGCACUGCGAGGAGUAGAAGCCA ................(((.......((((.....(((.....))).........(((((((((((.((.((((((((....)))))...))).)).)))))..))))))))))..))). (-28.26 = -28.60 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:26 2006