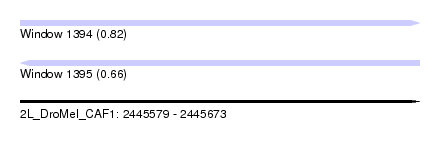
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,445,579 – 2,445,673 |
| Length | 94 |
| Max. P | 0.823641 |

| Location | 2,445,579 – 2,445,673 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -21.03 |
| Consensus MFE | -13.65 |
| Energy contribution | -14.60 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
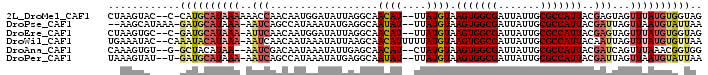
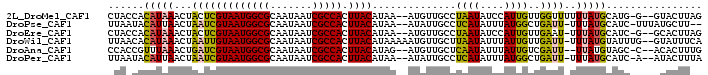
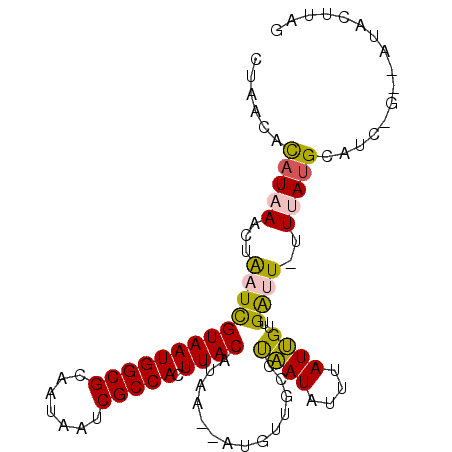
>2L_DroMel_CAF1 2445579 94 + 22407834 CUAAGUAC--C-CAUGCAUAAAAAACCAACAAUGGAUAUUAGGCAACAU--UUAUGUAAGUGGCGAUUAUUGCGCCAUUACGAGUAGUUUAUGUGGUAG .....(((--(-((((((((((...(((....)))......(....).)--)))))))(((((((.......)))))))............)).)))). ( -23.10) >DroPse_CAF1 26622 93 + 1 --AAGCAUAAA-GAUGCAUAAA-AAUCAGCCAUAAAUAUGAGGCAAUAU--UUAUGUAAGUGGCGAUUAUUGCGCCAUUACGAUUAGUUAAUGUAUUAA --.........-(((((((.((-((((...(((((((((......))))--)))))..(((((((.......)))))))..))))..)).))))))).. ( -22.60) >DroEre_CAF1 26640 93 + 1 CUAAGUGC--C-GAUGCAUAAA-AUUCAACAAUGGAUAUUAGGCAACAU--UUAUGUAAGUGGCGAUUAUUGCGCCAUUACGAGUAGUUUAUGUGGUAG .....(((--(-(.((((((((-(((((....)))))....(....).)--)))))))(((((((.......)))))))..............))))). ( -22.50) >DroWil_CAF1 39033 96 + 1 UGAAAUAC--CAAAUACAUAAA-AAUCAACAAUAAAUAUUAAGCAACAUUUUUAUGUAAGUGGCGAUUAUUGCGCCAUUACAAUUAGUUUAUGUGUUAA ........--..((((((((((-......................((((....)))).(((((((.......)))))))........)))))))))).. ( -18.40) >DroAna_CAF1 31971 92 + 1 CAAAGUGU--G-GCUACAUAA--AAUCGACAAUAAAUAUUGAGCAACAU--CUAUGUAAGUGGCGAUUAUUGCGCCAUUACGAUCAGUUUAAACGGUGG ((..((..--(-(((......--......((((....))))........--...(((((.(((((.......))))))))))...))))...))..)). ( -15.80) >DroPer_CAF1 26807 93 + 1 UAAAGUAU--U-GAUGCAUAAA-AAUCAGCCAUAAAUAUGAGGCAAUAU--UUAUGUAAGUGGCGAUUAUUGCGCCAUUACGAUUAGUUAAUGUAUUAA .......(--(-(((((((.((-((((...(((((((((......))))--)))))..(((((((.......)))))))..))))..)).))))))))) ( -23.80) >consensus CAAAGUAC__C_GAUGCAUAAA_AAUCAACAAUAAAUAUUAGGCAACAU__UUAUGUAAGUGGCGAUUAUUGCGCCAUUACGAUUAGUUUAUGUGGUAA ............((((((((((..(((..................((((....)))).(((((((.......)))))))..)))...)))))))))).. (-13.65 = -14.60 + 0.95)
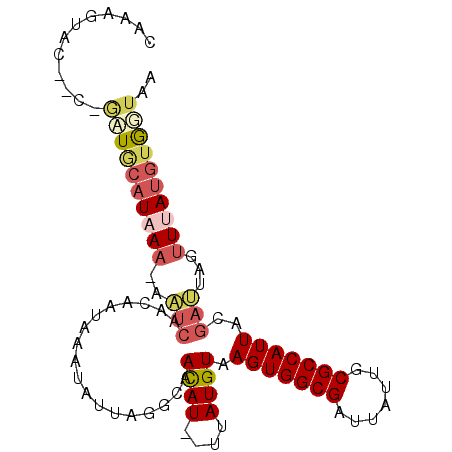
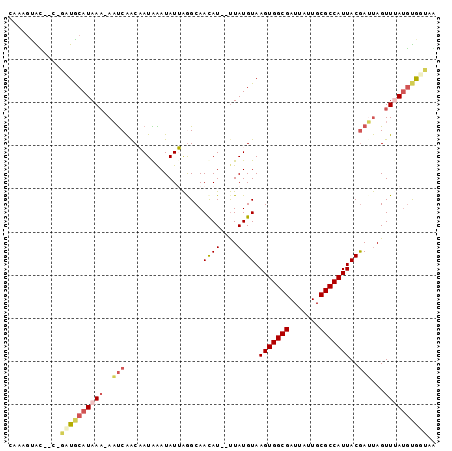
| Location | 2,445,579 – 2,445,673 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -16.85 |
| Consensus MFE | -11.09 |
| Energy contribution | -10.78 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2445579 94 - 22407834 CUACCACAUAAACUACUCGUAAUGGCGCAAUAAUCGCCACUUACAUAA--AUGUUGCCUAAUAUCCAUUGUUGGUUUUUUAUGCAUG-G--GUACUUAG .((((.(((.........(((((((((.......))))).))))....--.....((.(((...(((....)))....))).)))))-)--)))..... ( -20.10) >DroPse_CAF1 26622 93 - 1 UUAAUACAUUAACUAAUCGUAAUGGCGCAAUAAUCGCCACUUACAUAA--AUAUUGCCUCAUAUUUAUGGCUGAUU-UUUAUGCAUC-UUUAUGCUU-- ..................(((((((((.......))))).))))((((--(.((((((..........))).))).-)))))((((.-...))))..-- ( -17.10) >DroEre_CAF1 26640 93 - 1 CUACCACAUAAACUACUCGUAAUGGCGCAAUAAUCGCCACUUACAUAA--AUGUUGCCUAAUAUCCAUUGUUGAAU-UUUAUGCAUC-G--GCACUUAG ...((.............(((((((((.......))))).))))....--..(.(((.(((.((.((....)).))-.))).))).)-)--)....... ( -14.50) >DroWil_CAF1 39033 96 - 1 UUAACACAUAAACUAAUUGUAAUGGCGCAAUAAUCGCCACUUACAUAAAAAUGUUGCUUAAUAUUUAUUGUUGAUU-UUUAUGUAUUUG--GUAUUUCA ...............(((((......)))))....((((..(((((((((((...((............))..)))-))))))))..))--))...... ( -16.50) >DroAna_CAF1 31971 92 - 1 CCACCGUUUAAACUGAUCGUAAUGGCGCAAUAAUCGCCACUUACAUAG--AUGUUGCUCAAUAUUUAUUGUCGAUU--UUAUGUAGC-C--ACACUUUG .....(((......(((((((((((((.......))))).))))((((--(((((....)))))))))....))))--......)))-.--........ ( -17.40) >DroPer_CAF1 26807 93 - 1 UUAAUACAUUAACUAAUCGUAAUGGCGCAAUAAUCGCCACUUACAUAA--AUAUUGCCUCAUAUUUAUGGCUGAUU-UUUAUGCAUC-A--AUACUUUA ..................(((.((..(((..((((((((......(((--((((......))))))))))).))))-....)))..)-)--.))).... ( -15.50) >consensus CUAACACAUAAACUAAUCGUAAUGGCGCAAUAAUCGCCACUUACAUAA__AUGUUGCCUAAUAUUUAUUGUUGAUU_UUUAUGCAUC_G__AUACUUAG ......(((((...(((((((((((((.......))))).))))..............((((....))))..))))..)))))................ (-11.09 = -10.78 + -0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:21 2006