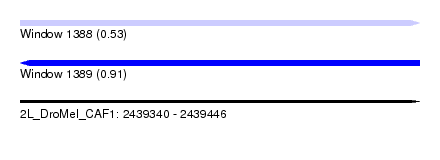
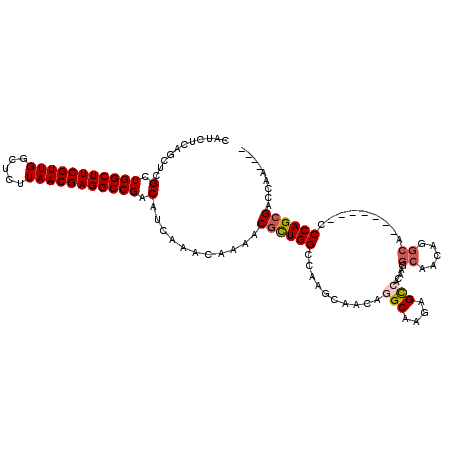
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,439,340 – 2,439,446 |
| Length | 106 |
| Max. P | 0.913935 |

| Location | 2,439,340 – 2,439,446 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -26.01 |
| Energy contribution | -26.23 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2439340 106 + 22407834 ----UUGGUCGCUGGG-------UGCCUGUUGCCUUGUGGCUCUUGCCUGUUGCUUGGCCAGCGUUUUGUUUGAUGUCGGCCUCGUUAAGAGCCAACGAAGCCAGCGAGCUGAGAUG ----..(.(((((((.-------(((.....)).((((((((((((......((..((((.(((((......))))).))))..)))))))))).))))).))))))).)....... ( -40.50) >DroSec_CAF1 18497 106 + 1 ----UUGGUCGCUGGG-------UGCCUGUUGCCUUGUGGCUCUUGCCUGCUGCUUGGCCAGCGUUUUGUUUGAUGUCGGCCUCGUUAAGAGCCAACGAAGCCAGCGAGCUGAGAUG ----..(.(((((((.-------(((.....)).((((((((((((......((..((((.(((((......))))).))))..)))))))))).))))).))))))).)....... ( -40.50) >DroSim_CAF1 18532 106 + 1 ----UUGGUCGCUGGG-------UGCCUGUUGCCUUGUGGCUCUUGCCUGCUGCUUGGCCAGCGUUUUGUUUGAUGUCGGCCUCGUUAAGAGCCAACGAAGCCAGCGAGCUGAGAUG ----..(.(((((((.-------(((.....)).((((((((((((......((..((((.(((((......))))).))))..)))))))))).))))).))))))).)....... ( -40.50) >DroEre_CAF1 20189 92 + 1 ----UUGGUCGCUGGC-------UGCCUGUUGC--------------CUGUUGCCUGGCCAGCGUUUUGUUUGAUGUCGGCCUCGUUAAGAGCCAACGAAGCCAGCGAGCUGAGAUG ----..(.((((((((-------(...(((((.--------------((.((((..((((.(((((......))))).))))..).))).)).))))).))))))))).)....... ( -40.00) >DroWil_CAF1 30367 89 + 1 GUUUCUGAUCUCUGGG-------UCUCUGUGGC--------------CUGU----GGUCCAACGUUUUGUUUGAUGUCGGCCUCGUUAAGAGCCAACGAAGCCAACGAGUAAAG--- ....((...(((..((-------..((..((((--------------((..----((.((.(((((......))))).))))......)).))))..))..))...)))...))--- ( -23.70) >DroYak_CAF1 19650 113 + 1 ----UUGGUCGCUGGCUGCUGGCUGCCUGUUGCCUUGUGACUCUGGCCUGUUGCUUGGCCAGCGUUUUGUUUGAUGUCGGCCUCGUUAAGAGCCAACGAAGCCAGCGAGCUGAGAUG ----..(.(((((((((..((((((((.((..(...)..))...)))((...((..((((.(((((......))))).))))..))..)))))))....))))))))).)....... ( -48.10) >consensus ____UUGGUCGCUGGG_______UGCCUGUUGCCUUGUGGCUCUUGCCUGUUGCUUGGCCAGCGUUUUGUUUGAUGUCGGCCUCGUUAAGAGCCAACGAAGCCAGCGAGCUGAGAUG ......(.(((((((.........................................((((.(((((......))))).))))(((((.......)))))..))))))).)....... (-26.01 = -26.23 + 0.22)

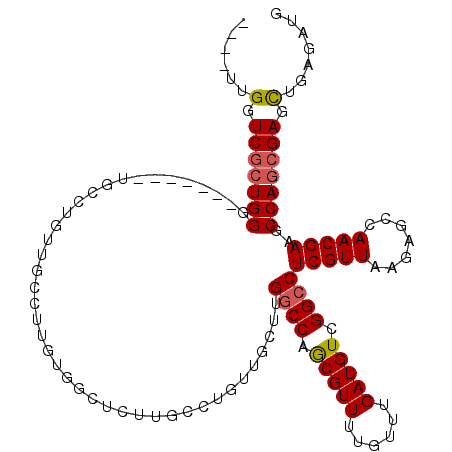
| Location | 2,439,340 – 2,439,446 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.04 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -24.25 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2439340 106 - 22407834 CAUCUCAGCUCGCUGGCUUCGUUGGCUCUUAACGAGGCCGACAUCAAACAAAACGCUGGCCAAGCAACAGGCAAGAGCCACAAGGCAACAGGCA-------CCCAGCGACCAA---- .......(.(((((((....(((((((........)))))))............(((.(((........)))....(((....)))....))).-------.))))))).)..---- ( -35.10) >DroSec_CAF1 18497 106 - 1 CAUCUCAGCUCGCUGGCUUCGUUGGCUCUUAACGAGGCCGACAUCAAACAAAACGCUGGCCAAGCAGCAGGCAAGAGCCACAAGGCAACAGGCA-------CCCAGCGACCAA---- .......(.(((((((....(((((((........)))))))............(((.(((........)))....(((....)))....))).-------.))))))).)..---- ( -35.10) >DroSim_CAF1 18532 106 - 1 CAUCUCAGCUCGCUGGCUUCGUUGGCUCUUAACGAGGCCGACAUCAAACAAAACGCUGGCCAAGCAGCAGGCAAGAGCCACAAGGCAACAGGCA-------CCCAGCGACCAA---- .......(.(((((((....(((((((........)))))))............(((.(((........)))....(((....)))....))).-------.))))))).)..---- ( -35.10) >DroEre_CAF1 20189 92 - 1 CAUCUCAGCUCGCUGGCUUCGUUGGCUCUUAACGAGGCCGACAUCAAACAAAACGCUGGCCAGGCAACAG--------------GCAACAGGCA-------GCCAGCGACCAA---- .......(.(((((((((..(((((((........)))))))............(((.(((.(....).)--------------))....))))-------)))))))).)..---- ( -38.20) >DroWil_CAF1 30367 89 - 1 ---CUUUACUCGUUGGCUUCGUUGGCUCUUAACGAGGCCGACAUCAAACAAAACGUUGGACC----ACAG--------------GCCACAGAGA-------CCCAGAGAUCAGAAAC ---.....((((((((((((((((.....)))))))))))))............((.((.((----...)--------------))))).))).-------................ ( -26.00) >DroYak_CAF1 19650 113 - 1 CAUCUCAGCUCGCUGGCUUCGUUGGCUCUUAACGAGGCCGACAUCAAACAAAACGCUGGCCAAGCAACAGGCCAGAGUCACAAGGCAACAGGCAGCCAGCAGCCAGCGACCAA---- .......(.(((((((((..(((((((........)))))))............((((((...((.....(((..........))).....)).))))))))))))))).)..---- ( -46.00) >consensus CAUCUCAGCUCGCUGGCUUCGUUGGCUCUUAACGAGGCCGACAUCAAACAAAACGCUGGCCAAGCAACAGGCAAGAGCCACAAGGCAACAGGCA_______CCCAGCGACCAA____ ...........(.(((((((((((.....))))))))))).)...........((((((..........(((....))).....((.....)).........))))))......... (-24.25 = -25.03 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:15 2006