| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,425,378 – 2,425,525 |
| Length | 147 |
| Max. P | 0.869007 |

| Location | 2,425,378 – 2,425,495 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.85 |
| Mean single sequence MFE | -21.64 |
| Consensus MFE | -17.11 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2425378 117 + 22407834 GAAGUUAUAGAAAUUGAUUUCUCUUUCAAGCAAG-UCUUUAAAAAUGCAUCUAACAAUGCCAUCAAUUUUAAAAGGUUUCCACAAAGCUCUUAGCUCCUCACAA-ACCUUUUCCGAUCU .........(((((((((....(((......)))-...........((((......)))).)))))))))(((((((((......(((.....)))......))-)))))))....... ( -16.50) >DroSec_CAF1 4873 117 + 1 GGGGUUAUAAAAAUUGAUUUCUCUUUCAAGCAAG-GCUUUGAAAAUGCAUCUAACAAUGCCAUCAAUUUUAAAAGGUUUCCACAAAGCUCUUAGCUCCACACAA-ACCUUUUCCGAUCU .(((((...(((((((((.....((((((((...-)).))))))..((((......)))).)))))))))(((((((((......(((.....)))......))-)))))))..))))) ( -23.40) >DroSim_CAF1 4894 117 + 1 GGGGUUAUAAAAAUUGAUUUCUCUUUCAAGCAGG-GCUUUGAAAAUGCAUCUAACAAUGCCAUCAAUUUUAAAAGGUUGCCACAAAGCUCUUAGCUCCACACAA-ACCUUUUCCGAUCU (((((((......((((........))))..(((-((((((.....((((......))))...((((((....))))))...))))))))))))))))......-.............. ( -25.10) >DroEre_CAF1 4893 118 + 1 GAGGUUAUAAAAAUUGAUUUCUCUUUUAAGCAAGAUGCCUGAAAAUGCAUCUAACAAUGCCAUCAAUUUUAAAAGGUUUCCACAAAGCUCUCAGCUCGCCGCAA-ACCUUUUCCGAUCU .(((((...(((((((((...........(((((((((.(....).)))))).....))).)))))))))(((((((((.(....(((.....)))....).))-)))))))..))))) ( -23.70) >DroYak_CAF1 4984 118 + 1 GGGGUUAUAAAAAUUGAUUUCUCUUUUAACCAAG-UCUUUGGAAAUGCAUCUAACAAUGCCAUCAAAUUUAAAAGGUUUUCACAAAGCUCUUAGCUCGCCACAAAACCUUUUCCGAUCU (..((((.......))))..).............-...((((((..((((......))))............((((((((.....(((.....)))......))))))))))))))... ( -19.50) >consensus GGGGUUAUAAAAAUUGAUUUCUCUUUCAAGCAAG_UCUUUGAAAAUGCAUCUAACAAUGCCAUCAAUUUUAAAAGGUUUCCACAAAGCUCUUAGCUCCACACAA_ACCUUUUCCGAUCU .(((((...(((((((((.....(((((((.......)))))))..((((......)))).)))))))))(((((((........(((.....))).........)))))))..))))) (-17.11 = -17.27 + 0.16)

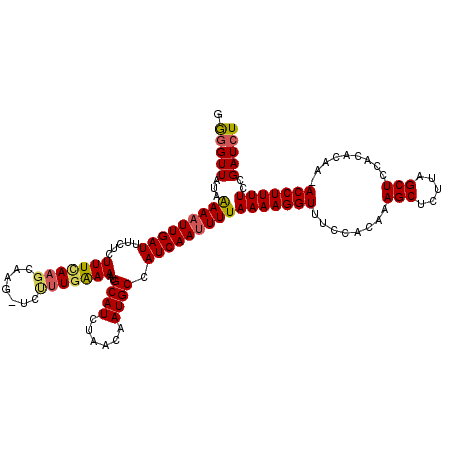
| Location | 2,425,378 – 2,425,495 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.85 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2425378 117 - 22407834 AGAUCGGAAAAGGU-UUGUGAGGAGCUAAGAGCUUUGUGGAAACCUUUUAAAAUUGAUGGCAUUGUUAGAUGCAUUUUUAAAGA-CUUGCUUGAAAGAGAAAUCAAUUUCUAUAACUUC .......(((((((-((.((..((((.....))))..)).))))))))).((((((((.(((((....)))))...........-....(((....)))..)))))))).......... ( -27.10) >DroSec_CAF1 4873 117 - 1 AGAUCGGAAAAGGU-UUGUGUGGAGCUAAGAGCUUUGUGGAAACCUUUUAAAAUUGAUGGCAUUGUUAGAUGCAUUUUCAAAGC-CUUGCUUGAAAGAGAAAUCAAUUUUUAUAACCCC .....(((((((((-((.((..((((.....))))..)).)))))))))(((((((((.(((((....))))).(((((((.((-...)))))))))....))))))))).....)).. ( -34.80) >DroSim_CAF1 4894 117 - 1 AGAUCGGAAAAGGU-UUGUGUGGAGCUAAGAGCUUUGUGGCAACCUUUUAAAAUUGAUGGCAUUGUUAGAUGCAUUUUCAAAGC-CCUGCUUGAAAGAGAAAUCAAUUUUUAUAACCCC .....(((((((((-(..((..((((.....))))..))..))))))))(((((((((.(((((....))))).(((((((.((-...)))))))))....))))))))).....)).. ( -30.80) >DroEre_CAF1 4893 118 - 1 AGAUCGGAAAAGGU-UUGCGGCGAGCUGAGAGCUUUGUGGAAACCUUUUAAAAUUGAUGGCAUUGUUAGAUGCAUUUUCAGGCAUCUUGCUUAAAAGAGAAAUCAAUUUUUAUAACCUC .....(((((((((-((.(.((((((.....)).)))).))))))))))(((((((((.(((((....))))).(((((((((.....))))....)))))))))))))).....)).. ( -31.90) >DroYak_CAF1 4984 118 - 1 AGAUCGGAAAAGGUUUUGUGGCGAGCUAAGAGCUUUGUGAAAACCUUUUAAAUUUGAUGGCAUUGUUAGAUGCAUUUCCAAAGA-CUUGGUUAAAAGAGAAAUCAAUUUUUAUAACCCC .....((((((((((((...((((((.....)).)))).))))))).............(((((....))))).))))).....-...((((((((((........))))).))))).. ( -24.20) >consensus AGAUCGGAAAAGGU_UUGUGGGGAGCUAAGAGCUUUGUGGAAACCUUUUAAAAUUGAUGGCAUUGUUAGAUGCAUUUUCAAAGA_CUUGCUUGAAAGAGAAAUCAAUUUUUAUAACCCC .....(((((((((.((.((..((((.....))))..)).)))))))))(((((((((.(((((....)))))................(((....)))..))))))))).....)).. (-22.04 = -22.52 + 0.48)

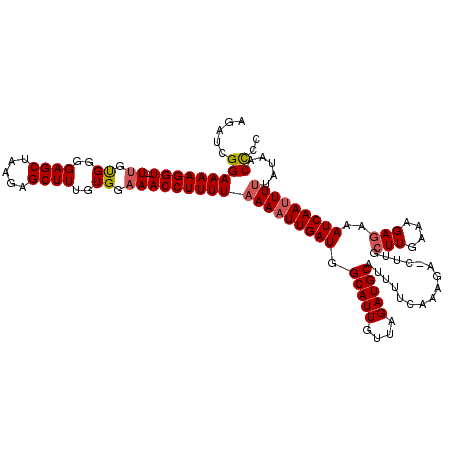
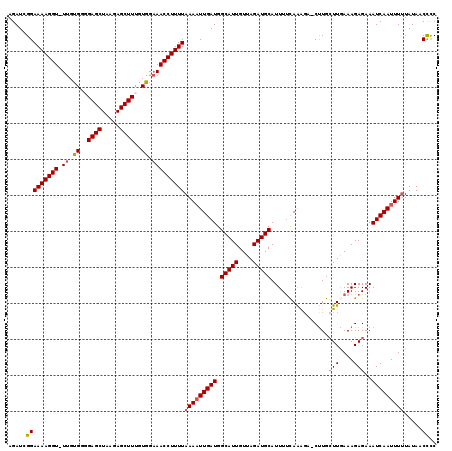
| Location | 2,425,417 – 2,425,525 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 95.20 |
| Mean single sequence MFE | -19.15 |
| Consensus MFE | -15.89 |
| Energy contribution | -16.09 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2425417 108 + 22407834 AAAAAUGCAUCUAACAAUGCCAUCAAUUUUAAAAGGUUUCCACAAAGCUCUUAGCUCCUCACAA-ACCUUUUCCGAUCUUGCAACACUCGAGUACCGAUGCUCAUCGCA .....(((.........(((.....(((..(((((((((......(((.....)))......))-)))))))..)))...)))......(((((....)))))...))) ( -18.80) >DroSec_CAF1 4912 108 + 1 GAAAAUGCAUCUAACAAUGCCAUCAAUUUUAAAAGGUUUCCACAAAGCUCUUAGCUCCACACAA-ACCUUUUCCGAUCUUGCAACACUCGAGUACCGAUGCUCAUCGCA .....(((.........(((.....(((..(((((((((......(((.....)))......))-)))))))..)))...)))......(((((....)))))...))) ( -18.80) >DroSim_CAF1 4933 108 + 1 GAAAAUGCAUCUAACAAUGCCAUCAAUUUUAAAAGGUUGCCACAAAGCUCUUAGCUCCACACAA-ACCUUUUCCGAUCUUGCAACACUCGAGUACCGAUGCUCAUCGCA .....(((.........(((.....(((..((((((((.......(((.....))).......)-)))))))..)))...)))......(((((....)))))...))) ( -18.84) >DroEre_CAF1 4933 108 + 1 GAAAAUGCAUCUAACAAUGCCAUCAAUUUUAAAAGGUUUCCACAAAGCUCUCAGCUCGCCGCAA-ACCUUUUCCGAUCUUGCAACACUUGAGUACCGAUGCUCAUCGCA .....(((.........(((.....(((..(((((((((.(....(((.....)))....).))-)))))))..)))...))).....((((((....))))))..))) ( -19.20) >DroYak_CAF1 5023 109 + 1 GGAAAUGCAUCUAACAAUGCCAUCAAAUUUAAAAGGUUUUCACAAAGCUCUUAGCUCGCCACAAAACCUUUUCCGAUCUUGCAACACUUGAGUACCGAUGCUCAUCGCA ((((..((((......))))............((((((((.....(((.....)))......)))))))))))).....(((......((((((....))))))..))) ( -20.10) >consensus GAAAAUGCAUCUAACAAUGCCAUCAAUUUUAAAAGGUUUCCACAAAGCUCUUAGCUCCACACAA_ACCUUUUCCGAUCUUGCAACACUCGAGUACCGAUGCUCAUCGCA .....(((.........(((.....(((..(((((((........(((.....))).........)))))))..)))...)))......(((((....)))))...))) (-15.89 = -16.09 + 0.20)

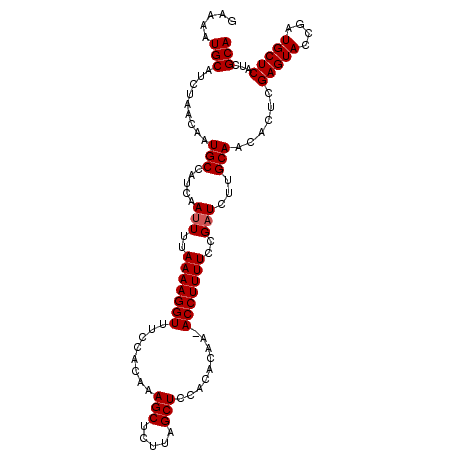
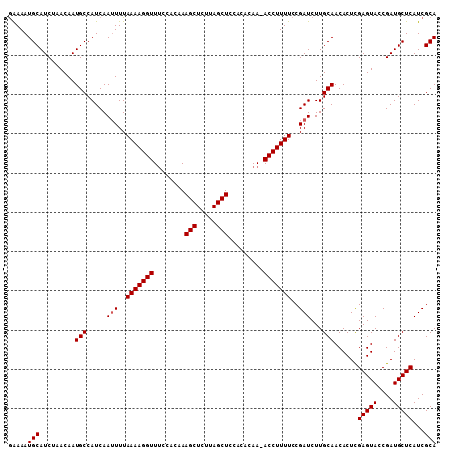
| Location | 2,425,417 – 2,425,525 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 95.20 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -24.30 |
| Energy contribution | -24.34 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2425417 108 - 22407834 UGCGAUGAGCAUCGGUACUCGAGUGUUGCAAGAUCGGAAAAGGU-UUGUGAGGAGCUAAGAGCUUUGUGGAAACCUUUUAAAAUUGAUGGCAUUGUUAGAUGCAUUUUU .((((((..(((((((..((((...........))))(((((((-((.((..((((.....))))..)).)))))))))...))))))).))))))............. ( -28.60) >DroSec_CAF1 4912 108 - 1 UGCGAUGAGCAUCGGUACUCGAGUGUUGCAAGAUCGGAAAAGGU-UUGUGUGGAGCUAAGAGCUUUGUGGAAACCUUUUAAAAUUGAUGGCAUUGUUAGAUGCAUUUUC .((((((..(((((((..((((...........))))(((((((-((.((..((((.....))))..)).)))))))))...))))))).))))))............. ( -31.60) >DroSim_CAF1 4933 108 - 1 UGCGAUGAGCAUCGGUACUCGAGUGUUGCAAGAUCGGAAAAGGU-UUGUGUGGAGCUAAGAGCUUUGUGGCAACCUUUUAAAAUUGAUGGCAUUGUUAGAUGCAUUUUC .((((((..(((((((....(((.(((((.(((((......)))-))..(..((((.....))))..).)))))))).....))))))).))))))............. ( -28.50) >DroEre_CAF1 4933 108 - 1 UGCGAUGAGCAUCGGUACUCAAGUGUUGCAAGAUCGGAAAAGGU-UUGCGGCGAGCUGAGAGCUUUGUGGAAACCUUUUAAAAUUGAUGGCAUUGUUAGAUGCAUUUUC ........(((((...((...((((((((((......(((((((-((.(.((((((.....)).)))).))))))))))....))).)))))))))..)))))...... ( -28.70) >DroYak_CAF1 5023 109 - 1 UGCGAUGAGCAUCGGUACUCAAGUGUUGCAAGAUCGGAAAAGGUUUUGUGGCGAGCUAAGAGCUUUGUGAAAACCUUUUAAAUUUGAUGGCAUUGUUAGAUGCAUUUCC ........((((((((..(((...((..(((((((......)))))))..))((((.....))))..)))..))).........((((......)))))))))...... ( -28.00) >consensus UGCGAUGAGCAUCGGUACUCGAGUGUUGCAAGAUCGGAAAAGGU_UUGUGGGGAGCUAAGAGCUUUGUGGAAACCUUUUAAAAUUGAUGGCAUUGUUAGAUGCAUUUUC ........(((((...((...((((((((((......(((((((.((.((..((((.....))))..)).)))))))))....))).)))))))))..)))))...... (-24.30 = -24.34 + 0.04)


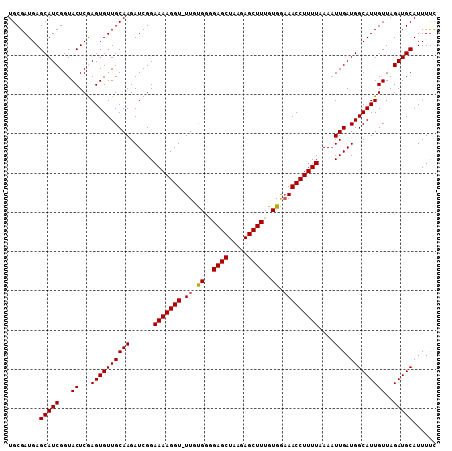
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:12 2006