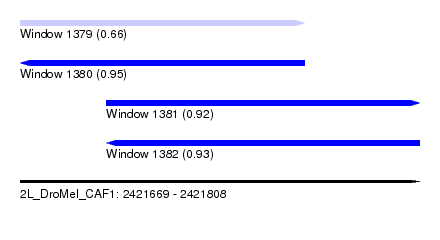
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,421,669 – 2,421,808 |
| Length | 139 |
| Max. P | 0.951598 |

| Location | 2,421,669 – 2,421,768 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -24.13 |
| Energy contribution | -25.88 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2421669 99 + 22407834 CGGGUCUUUGAGCAGCCGGAGGAGCAGAAGAACACAUCGCUGCCCGAGCACAAGGUUUUUGUGGUGGGCAGCAAUGUGAUAGCCACUUCCACAGCGGCU .(..((((((......))))))..).......(((((.((((((((..((((((...)))))).)))))))).)))))..((((.((.....)).)))) ( -37.00) >DroPse_CAF1 828 99 + 1 AAUGUCUUUGAGGUGUCGGACAAGCAAGACAACCCUUUGCUACCCAAGCACAAAGUCUAUGUGGUGGGUAGCAAUGCCAUAGCUACCUCAACAGCAGCC ..(((..((((((((((..........)))......((((((((((..((((.......)))).))))))))))..........)))))))..)))... ( -34.20) >DroSim_CAF1 775 99 + 1 CGGGUCUUUGAGCAGCCGGAGGAGCAGAAGAACACAUCUCUGCCUGAGCACAAGGUCUUUGUAGUGGGCAGCAAUGUGAUUGCCACUUCCACAGCGGCU ..........(((.((.(((((.((((.....(((((..(((((((...(((((...)))))..)))))))..))))).))))..)))))...)).))) ( -29.10) >DroEre_CAF1 775 99 + 1 CGGGUGUUUGAGCAGCCGGAGGAGCAGAUGCACACAUCGCUGCCCGAGCACAAGGUAUUUGUGGUGGGCAGCAAUCUGAUAGCCACUUCCACAGCGGCC .((.(((....))).))(((((.((.((((....))))((((((((..((((((...)))))).)))))))).........))..)))))......... ( -39.00) >DroYak_CAF1 775 99 + 1 CGGGUGUUUGAGCAGCCGGAGGAGCAGAGCAACACAUCGCUGCCCGAGCACAAGGUCUUUGUGGUGGGCAGCAAUGUGAUAGCCACUUCCACAGCGGCC .((.(((....))).))(((((.((...))..(((((.((((((((..((((((...)))))).)))))))).))))).......)))))......... ( -40.00) >DroPer_CAF1 828 99 + 1 AAUGUCUUUGAGGUGUCGGACAAGCAAGACAACCCUUUGCUACCCAAGCACAAAGUCUAUGUGGUGGGUAGCAAUGCCAUAGCUACCUCAACGGCAGCC ..((((.((((((((((..........)))......((((((((((..((((.......)))).))))))))))..........))))))).))))... ( -36.60) >consensus CGGGUCUUUGAGCAGCCGGAGGAGCAGAACAACACAUCGCUGCCCGAGCACAAGGUCUUUGUGGUGGGCAGCAAUGUGAUAGCCACUUCCACAGCGGCC (((.(((....))).)))..............(((((.((((((((..((((((...)))))).)))))))).)))))...(((.((.....)).))). (-24.13 = -25.88 + 1.75)

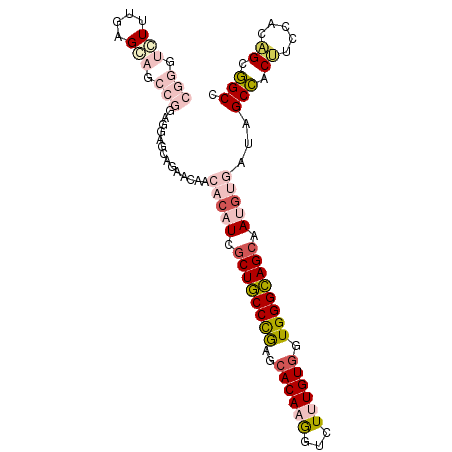
| Location | 2,421,669 – 2,421,768 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -39.05 |
| Consensus MFE | -31.95 |
| Energy contribution | -32.18 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2421669 99 - 22407834 AGCCGCUGUGGAAGUGGCUAUCACAUUGCUGCCCACCACAAAAACCUUGUGCUCGGGCAGCGAUGUGUUCUUCUGCUCCUCCGGCUGCUCAAAGACCCG (((.((((..(.((..(....(((((((((((((..(((((.....)))))...))))))))))))).....)..)).)..)))).))).......... ( -44.10) >DroPse_CAF1 828 99 - 1 GGCUGCUGUUGAGGUAGCUAUGGCAUUGCUACCCACCACAUAGACUUUGUGCUUGGGUAGCAAAGGGUUGUCUUGCUUGUCCGACACCUCAAAGACAUU (((((((.....)))))))(((...((((((((((.((((.......))))..)))))))))).(((.((((..........)))).))).....))). ( -34.70) >DroSim_CAF1 775 99 - 1 AGCCGCUGUGGAAGUGGCAAUCACAUUGCUGCCCACUACAAAGACCUUGUGCUCAGGCAGAGAUGUGUUCUUCUGCUCCUCCGGCUGCUCAAAGACCCG (((.((((..(.((..(....(((((..(((((...(((((.....)))))....)))))..))))).....)..)).)..)))).))).......... ( -32.80) >DroEre_CAF1 775 99 - 1 GGCCGCUGUGGAAGUGGCUAUCAGAUUGCUGCCCACCACAAAUACCUUGUGCUCGGGCAGCGAUGUGUGCAUCUGCUCCUCCGGCUGCUCAAACACCCG (((.((((..(.((..(....((.((((((((((..(((((.....)))))...)))))))))).)).....)..)).)..)))).))).......... ( -40.20) >DroYak_CAF1 775 99 - 1 GGCCGCUGUGGAAGUGGCUAUCACAUUGCUGCCCACCACAAAGACCUUGUGCUCGGGCAGCGAUGUGUUGCUCUGCUCCUCCGGCUGCUCAAACACCCG (((.((((..(.((((((...(((((((((((((..(((((.....)))))...)))))))))))))..)))..))).)..)))).))).......... ( -45.20) >DroPer_CAF1 828 99 - 1 GGCUGCCGUUGAGGUAGCUAUGGCAUUGCUACCCACCACAUAGACUUUGUGCUUGGGUAGCAAAGGGUUGUCUUGCUUGUCCGACACCUCAAAGACAUU (((((((.....)))))))(((...((((((((((.((((.......))))..)))))))))).(((.((((..........)))).))).....))). ( -37.30) >consensus GGCCGCUGUGGAAGUGGCUAUCACAUUGCUGCCCACCACAAAGACCUUGUGCUCGGGCAGCGAUGUGUUCUUCUGCUCCUCCGGCUGCUCAAAGACCCG (((.((((.(((.((((....(((((((((((((..(((((.....)))))...))))))))))))).....)))))))..)))).))).......... (-31.95 = -32.18 + 0.23)


| Location | 2,421,699 – 2,421,808 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.94 |
| Mean single sequence MFE | -40.84 |
| Consensus MFE | -30.51 |
| Energy contribution | -30.82 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2421699 109 + 22407834 AACACAUCGCUGCCCGAGCACAAGGUUUUUGUGGUGGGCAGCAAUGUGAUAGCCACUUCCACAGCGGCUCAUGAGGCCGAAAGGCUGGGCUACAUUCCGUGCGUGCUCA ..(((((.((((((((..((((((...)))))).)))))))).)))))..((((.((.....)).)))).....((((....))))((((..(((...)))...)))). ( -46.00) >DroPse_CAF1 858 109 + 1 AACCCUUUGCUACCCAAGCACAAAGUCUAUGUGGUGGGUAGCAAUGCCAUAGCUACCUCAACAGCAGCCCAACAGGCUGCCAAACUAGGAUACAUUCCCUGCGUCCUAA ......((((((((((..((((.......)))).))))))))))...................((((((.....)))))).....(((((..((.....))..))))). ( -37.70) >DroSim_CAF1 805 109 + 1 AACACAUCUCUGCCUGAGCACAAGGUCUUUGUAGUGGGCAGCAAUGUGAUUGCCACUUCCACAGCGGCUCAUGAGGCCGAAAAGCUGGGCUACAUUCCCUGCGUGCUCA ...............((((((..((((((....((((((.((..((((...........)))))).)))))))))))).....((.(((.......))).)))))))). ( -36.90) >DroEre_CAF1 805 109 + 1 CACACAUCGCUGCCCGAGCACAAGGUAUUUGUGGUGGGCAGCAAUCUGAUAGCCACUUCCACAGCGGCCAAGGAGGCCGAAAAUCUGGGUUAUAUUCCCUGCGUGCUCA (((.((..((((((((..((((((...)))))).))))))))......((((((..........(((((.....)))))........))))))......)).))).... ( -39.67) >DroYak_CAF1 805 109 + 1 AACACAUCGCUGCCCGAGCACAAGGUCUUUGUGGUGGGCAGCAAUGUGAUAGCCACUUCCACAGCGGCCAAGGAGGCCGAAAAUCUGGGUUACAUUCCCUGCGUGCUCA ..(((((.((((((((..((((((...)))))).)))))))).)))))..(((.((........(((((.....))))).......(((.......)))...))))).. ( -42.80) >DroPer_CAF1 858 109 + 1 AACCCUUUGCUACCCAAGCACAAAGUCUAUGUGGUGGGUAGCAAUGCCAUAGCUACCUCAACGGCAGCCCAACAGGCUGCCAAACUAGGAUACAUUCCCUGCGUCCUAA ......((((((((((..((((.......)))).))))))))))..................(((((((.....)))))))....(((((..((.....))..))))). ( -42.00) >consensus AACACAUCGCUGCCCGAGCACAAGGUCUUUGUGGUGGGCAGCAAUGUGAUAGCCACUUCCACAGCGGCCCAAGAGGCCGAAAAACUGGGAUACAUUCCCUGCGUGCUCA ..(((((.((((((((..((((((...)))))).)))))))).))))).(((((..........(((((.....)))))........)))))................. (-30.51 = -30.82 + 0.31)

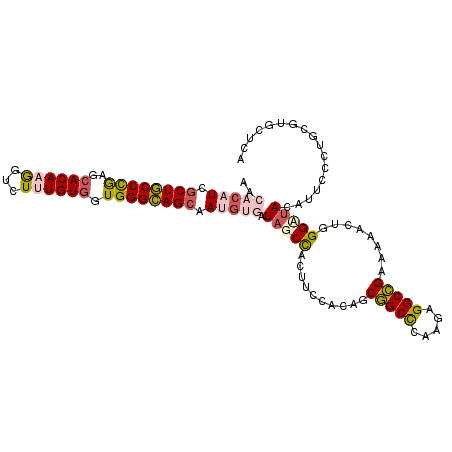
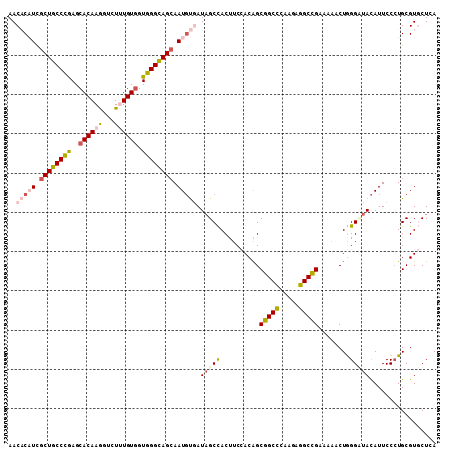
| Location | 2,421,699 – 2,421,808 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.94 |
| Mean single sequence MFE | -44.68 |
| Consensus MFE | -34.98 |
| Energy contribution | -34.68 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2421699 109 - 22407834 UGAGCACGCACGGAAUGUAGCCCAGCCUUUCGGCCUCAUGAGCCGCUGUGGAAGUGGCUAUCACAUUGCUGCCCACCACAAAAACCUUGUGCUCGGGCAGCGAUGUGUU ((((...((.(((((.((......)).)))))))))))..(((((((.....)))))))..(((((((((((((..(((((.....)))))...))))))))))))).. ( -48.60) >DroPse_CAF1 858 109 - 1 UUAGGACGCAGGGAAUGUAUCCUAGUUUGGCAGCCUGUUGGGCUGCUGUUGAGGUAGCUAUGGCAUUGCUACCCACCACAUAGACUUUGUGCUUGGGUAGCAAAGGGUU ((((((..((.....))..))))))...(((((((.....))))))).........((....)).((((((((((.((((.......))))..))))))))))...... ( -43.10) >DroSim_CAF1 805 109 - 1 UGAGCACGCAGGGAAUGUAGCCCAGCUUUUCGGCCUCAUGAGCCGCUGUGGAAGUGGCAAUCACAUUGCUGCCCACUACAAAGACCUUGUGCUCAGGCAGAGAUGUGUU ((((((((.(((...(((((((((((.....((((....).))))))).))..(..(((((...)))))..)...)))))....))))))))))).(((....)))... ( -38.30) >DroEre_CAF1 805 109 - 1 UGAGCACGCAGGGAAUAUAACCCAGAUUUUCGGCCUCCUUGGCCGCUGUGGAAGUGGCUAUCAGAUUGCUGCCCACCACAAAUACCUUGUGCUCGGGCAGCGAUGUGUG ..(((.(((.(((.......))).......(((((.....)))))........))))))..((.((((((((((..(((((.....)))))...)))))))))).)).. ( -44.30) >DroYak_CAF1 805 109 - 1 UGAGCACGCAGGGAAUGUAACCCAGAUUUUCGGCCUCCUUGGCCGCUGUGGAAGUGGCUAUCACAUUGCUGCCCACCACAAAGACCUUGUGCUCGGGCAGCGAUGUGUU ..(((.(((.(((.......))).......(((((.....)))))........))))))..(((((((((((((..(((((.....)))))...))))))))))))).. ( -48.20) >DroPer_CAF1 858 109 - 1 UUAGGACGCAGGGAAUGUAUCCUAGUUUGGCAGCCUGUUGGGCUGCCGUUGAGGUAGCUAUGGCAUUGCUACCCACCACAUAGACUUUGUGCUUGGGUAGCAAAGGGUU ((((((..((.....))..))))))...(((((((.....))))))).........((....)).((((((((((.((((.......))))..))))))))))...... ( -45.60) >consensus UGAGCACGCAGGGAAUGUAACCCAGAUUUUCGGCCUCAUGGGCCGCUGUGGAAGUGGCUAUCACAUUGCUGCCCACCACAAAGACCUUGUGCUCGGGCAGCGAUGUGUU .(((......(((.......))).(((....))))))...(((((((.....)))))))..(((((((((((((..(((((.....)))))...))))))))))))).. (-34.98 = -34.68 + -0.30)

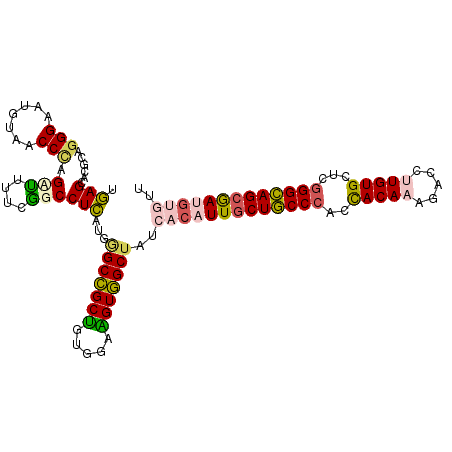
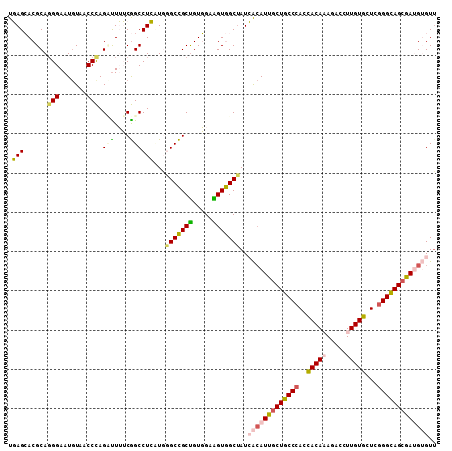
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:08 2006