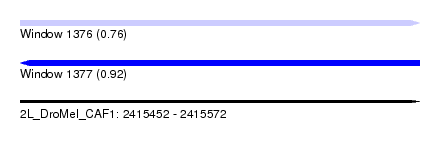
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,415,452 – 2,415,572 |
| Length | 120 |
| Max. P | 0.922590 |

| Location | 2,415,452 – 2,415,572 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.56 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
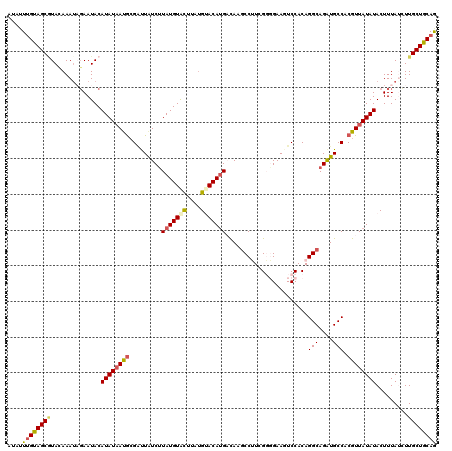
>2L_DroMel_CAF1 2415452 120 + 22407834 AUAUUUGUAGCUUAUAAAUAGAAUACAUAUUAUACGAUUAUCCUAUGUGCUUAUGUACAAGACAAGCCUUGGGGGAAAUCCACAGGCAGGUGCCACGUUAUAUACUUUAUCUUGCUGCAG .....((((((......(((((....((((...(((.....(((.(((((....)))))......((((((((.....)))).))))))).....))).))))..)))))...)))))). ( -27.30) >DroSec_CAF1 4299 117 + 1 A---UUGUAGCGUACAAAUAGAAUACAUAUAAUGCGAUUAUCUUAUGUACUUAUGUACAUGACAAGCCUUCGGGGAAGUCCACAGGCAGAUGCCACGUUAUAUACUUUAGUUUGCUGCAG .---.(((((((.((.....(....)((((((((.((((.((((((((((....))))))))....((....))))))))....(((....))).))))))))......)).))))))). ( -31.60) >DroSim_CAF1 3973 120 + 1 AUAUUUGUAGCGUACAAAUAGAAUACAUAUAAUGCGAUUAUCUUAUGUACUUAUGUACAUGACAAGCCUUCGGGGAAGUCCACAGGCAGAUGCCACGUUAUAUACUUUAGCUUGCUGCAG .....(((((((((.........)))((((((((.((((.((((((((((....))))))))....((....))))))))....(((....))).))))))))..........)))))). ( -31.30) >DroEre_CAF1 4134 118 + 1 AUAUUCGCAGCGUACAAAACUAAUACAUAUAACGCGUCUAUCUUAUGUAUU--UGCACAUGUCAUUCCCUGGCUGAAUUCCACAGGCAGAUGCCACGUUAUAUACUUUAUCUUGCUGCAA ......((((((((.........)))(((((((((((((....(((((...--...)))))......((((...........)))).))))))...)))))))..........))))).. ( -26.70) >DroYak_CAF1 4750 118 + 1 AUAUUUGCAGCAUAUAAAUCGAAUACAUAUAACGGGGUUGUCUUAUGUACU--CGAACAUGUUAGGCCGUAGCUGAAUUCCACAGGCAGAUGCCACGUUAUAUACUUUAUCUUGCUGCAA ....((((((((.(((((..(....)((((((((.((((....(((((...--...)))))...))))................(((....))).))))))))..)))))..)))))))) ( -30.30) >consensus AUAUUUGUAGCGUACAAAUAGAAUACAUAUAAUGCGAUUAUCUUAUGUACUUAUGUACAUGACAAGCCUUCGGGGAAGUCCACAGGCAGAUGCCACGUUAUAUACUUUAUCUUGCUGCAG ....((((((((..............((((((((.........(((((((....))))))).......................(((....))).)))))))).........)))))))) (-20.00 = -20.32 + 0.32)

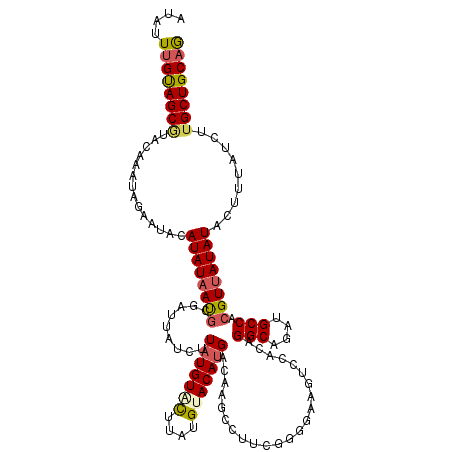
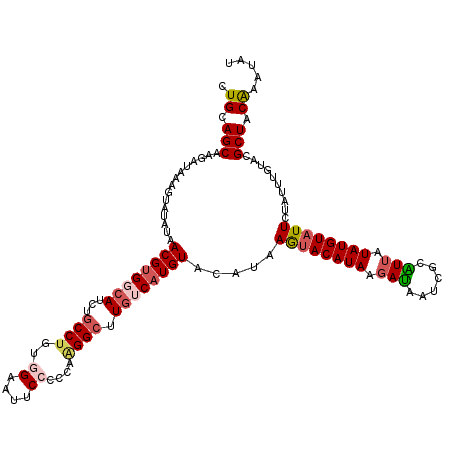
| Location | 2,415,452 – 2,415,572 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.56 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.76 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2415452 120 - 22407834 CUGCAGCAAGAUAAAGUAUAUAACGUGGCACCUGCCUGUGGAUUUCCCCCAAGGCUUGUCUUGUACAUAAGCACAUAGGAUAAUCGUAUAAUAUGUAUUCUAUUUAUAAGCUACAAAUAU .((.(((.(((((.(((((((((((.(((....)))))).((((..((.....((((((.......)))))).....))..))))......)))))))).)))))....))).))..... ( -24.90) >DroSec_CAF1 4299 117 - 1 CUGCAGCAAACUAAAGUAUAUAACGUGGCAUCUGCCUGUGGACUUCCCCGAAGGCUUGUCAUGUACAUAAGUACAUAAGAUAAUCGCAUUAUAUGUAUUCUAUUUGUACGCUACAA---U .((.(((.................((((((...((((.(((......))).)))).))))))(((((..((((((((.(((......))).)))))))).....)))))))).)).---. ( -31.00) >DroSim_CAF1 3973 120 - 1 CUGCAGCAAGCUAAAGUAUAUAACGUGGCAUCUGCCUGUGGACUUCCCCGAAGGCUUGUCAUGUACAUAAGUACAUAAGAUAAUCGCAUUAUAUGUAUUCUAUUUGUACGCUACAAAUAU .((.(((.................((((((...((((.(((......))).)))).))))))(((((..((((((((.(((......))).)))))))).....)))))))).))..... ( -31.00) >DroEre_CAF1 4134 118 - 1 UUGCAGCAAGAUAAAGUAUAUAACGUGGCAUCUGCCUGUGGAAUUCAGCCAGGGAAUGACAUGUGCA--AAUACAUAAGAUAGACGCGUUAUAUGUAUUAGUUUUGUACGCUGCGAAUAU (((((((..........((((((((((..((((..((.(((.......))).))......(((((..--..))))).))))...))))))))))((((.......))))))))))).... ( -33.20) >DroYak_CAF1 4750 118 - 1 UUGCAGCAAGAUAAAGUAUAUAACGUGGCAUCUGCCUGUGGAAUUCAGCUACGGCCUAACAUGUUCG--AGUACAUAAGACAACCCCGUUAUAUGUAUUCGAUUUAUAUGCUGCAAAUAU ((((((((..(((((.......(((((......(((.((((.......)))))))....)))))(((--((((((((.(((......))).)))))))))))))))).)))))))).... ( -39.50) >consensus CUGCAGCAAGAUAAAGUAUAUAACGUGGCAUCUGCCUGUGGAAUUCCCCCAAGGCUUGUCAUGUACAUAAGUACAUAAGAUAAUCGCAUUAUAUGUAUUCUAUUUGUACGCUACAAAUAU .((.(((...............((((((((...((((..((....))....)))).)))))))).....((((((((.(((......))).))))))))..........))).))..... (-19.60 = -20.76 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:04 2006