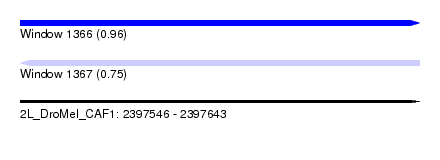
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,397,546 – 2,397,643 |
| Length | 97 |
| Max. P | 0.956281 |

| Location | 2,397,546 – 2,397,643 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -18.82 |
| Energy contribution | -20.38 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
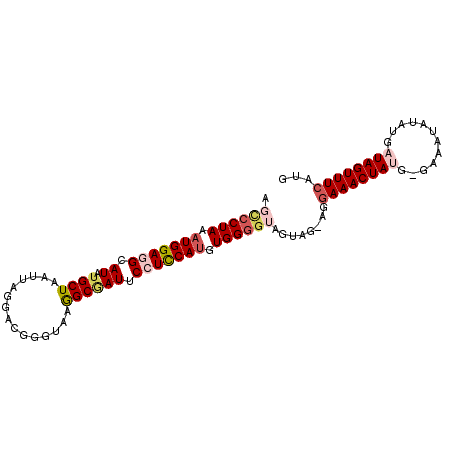
>2L_DroMel_CAF1 2397546 97 + 22407834 AGUCCUAAAUGGAGGCAUAUGCUAAUUAGGAUGGGUAAGGCGAUUCCUCCAAGUGGGGUAGUAGUAGGAAACUAUG-GAAAUAUAUGAUAGUUUCAUG ..(((((..((((((.((.((((...............)))))).))))))..))))).....((((....)))).-(((((........)))))... ( -25.16) >DroSec_CAF1 15505 96 + 1 AGCCCUAAAUGGAGGCAUAUGCUAAUUAGGACGGGCAAGGCGAUUCCUCCAUGUGGUGUAGUAA-ACGAAACUAUG-GAAAUAUAUGAUAGUUUCAUG .((((...(((((((.((.((((...............)))))).)))))))..)).)).....-..((((((((.-(.......).))))))))... ( -24.06) >DroSim_CAF1 16264 92 + 1 AGCCCUAAAUGGAGGCAUAUGCUAAUUAGGACGGGCAAGGCGAUUCCUCCAUGUGGGGUAGU-----GAAACUAUG-GAGAUAUAUGAUAGUUUCAUG .((((((.(((((((.((.((((...............)))))).))))))).)))))).((-----((((((((.-(.......).)))))))))). ( -32.46) >DroEre_CAF1 16661 95 + 1 --CCCUAAAUGGAGGCAUAUGCUAAUUAGAACGGGUAAAGCAAUCCGUCCAUGUGGGGCGGCAGAGAGAAACUACA-AAAAUAUAUGAUAGUUUCACA --(((((.((((((((....)))........(((((......)))))))))).))))).........(((((((((-........)).)))))))... ( -27.10) >DroYak_CAF1 16456 96 + 1 --GCCUAAAUGGAGGCAUAUGCUAAUUAGAACGGGUAAAGCCAUCCGUUCAUGUGGGGUAGUGGAAGAAAACUACGGAAAAUAUAUGAUAGUUUCAUG --((((......))))((((.((.....((((((((......)))))))).......(((((........)))))))...))))((((.....)))). ( -24.20) >consensus AGCCCUAAAUGGAGGCAUAUGCUAAUUAGGACGGGUAAGGCGAUUCCUCCAUGUGGGGUAGUAG_AGGAAACUAUG_GAAAUAUAUGAUAGUUUCAUG .((((((.(((((((.((.((((...............)))))).))))))).))))))........((((((((............))))))))... (-18.82 = -20.38 + 1.56)

| Location | 2,397,546 – 2,397,643 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -18.92 |
| Consensus MFE | -12.53 |
| Energy contribution | -12.93 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
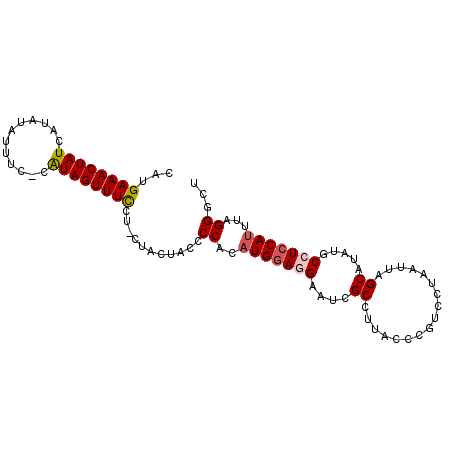
>2L_DroMel_CAF1 2397546 97 - 22407834 CAUGAAACUAUCAUAUAUUUC-CAUAGUUUCCUACUACUACCCCACUUGGAGGAAUCGCCUUACCCAUCCUAAUUAGCAUAUGCCUCCAUUUAGGACU ...((((((((..........-.))))))))..........((....((((((....((.................)).....))))))....))... ( -16.33) >DroSec_CAF1 15505 96 - 1 CAUGAAACUAUCAUAUAUUUC-CAUAGUUUCGU-UUACUACACCACAUGGAGGAAUCGCCUUGCCCGUCCUAAUUAGCAUAUGCCUCCAUUUAGGGCU .((((((((((..........-.))))))))))-........((..(((((((........(((............)))....)))))))...))... ( -19.70) >DroSim_CAF1 16264 92 - 1 CAUGAAACUAUCAUAUAUCUC-CAUAGUUUC-----ACUACCCCACAUGGAGGAAUCGCCUUGCCCGUCCUAAUUAGCAUAUGCCUCCAUUUAGGGCU ..(((((((((..........-.))))))))-----)...(((...(((((((........(((............)))....)))))))...))).. ( -22.10) >DroEre_CAF1 16661 95 - 1 UGUGAAACUAUCAUAUAUUUU-UGUAGUUUCUCUCUGCCGCCCCACAUGGACGGAUUGCUUUACCCGUUCUAAUUAGCAUAUGCCUCCAUUUAGGG-- .(.(((((((.((........-))))))))).).......(((...(((((.((..((((...............))))....)))))))...)))-- ( -21.46) >DroYak_CAF1 16456 96 - 1 CAUGAAACUAUCAUAUAUUUUCCGUAGUUUUCUUCCACUACCCCACAUGAACGGAUGGCUUUACCCGUUCUAAUUAGCAUAUGCCUCCAUUUAGGC-- .((((.....)))).........(((((........))))).......((((((..........))))))............((((......))))-- ( -15.00) >consensus CAUGAAACUAUCAUAUAUUUC_CAUAGUUUCCU_CUACUACCCCACAUGGAGGAAUCGCCUUACCCGUCCUAAUUAGCAUAUGCCUCCAUUUAGGGCU ...((((((((............))))))))...........((..(((((((....((.................)).....)))))))...))... (-12.53 = -12.93 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:53 2006