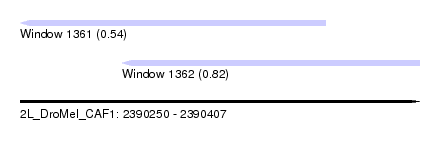
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,390,250 – 2,390,407 |
| Length | 157 |
| Max. P | 0.821763 |

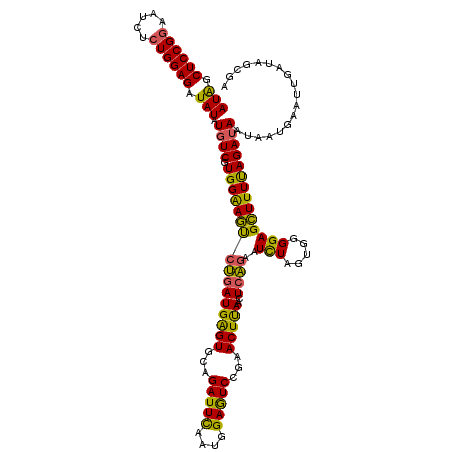
| Location | 2,390,250 – 2,390,370 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -27.88 |
| Energy contribution | -26.04 |
| Covariance contribution | -1.84 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2390250 120 - 22407834 AUAGCUCCGGAAUAUCUGGAGGUAUAUGUCGUGGAAGUCUGAUGGGUGCAGAUUCAAUGGAGUCCGAACUUAACUCAGAAUCUAGUGGGGAGCUUUUAGAUAAUAAUGAAUUGAUUGCAA (((.((((((.....)))))).))).(((.((.((..((((((((((.(.(((((....))))).).)))))..)))))((((((.((....)).)))))).........)).)).))). ( -32.30) >DroSec_CAF1 8253 120 - 1 AUGGCUCCGGAAUCUCUGGAGAUAUAUGUCUUGGAAGUCUGAUGAGUGCAGAUUCAUUGGAGUCCGAACUUAACUCAGAAUCUAGUGGGGAGCUUUUAGAUAACAAUAAAUUGGUAGCGA ...(((((((....(((((((((....)))))(((..((..((((((....))))))..)).)))..........))))((((((.((....)).)))))).........)))).))).. ( -32.90) >DroSim_CAF1 8934 120 - 1 AUAGCUCCGGAAUCUCUGGAGAUAUAUGUCUUGGAAGUCUGAUGAGUGCAGAUUUAUUGGAGUCCGAACUUAACUCACAAUCUAGUGGGGAGCUUUUAGAUAACAAUGAAUUGGUAGCGA (((.((((((.....)))))).)))...(((..(....)..).)).(((.(((((((((..(((..((.....(((((......))))).....))..)))..))))))))).))).... ( -30.50) >DroEre_CAF1 9167 120 - 1 AUAGCUCCGGAAUCUAUGGAGGUAUAUGUCGUGGGAGCCGGAUGAGUGCAGAUUCAAUGGAAUCGGAACUCAAUUCUGAUUUUAGUUCGGAGCUUUUAGAUAUUAGUGAAGUGAUCGCGA (((.(((((.......))))).)))(((((.(((((((....(((((...(((((....)))))...)))))..(((((.......)))))))))))))))))..((((.....)))).. ( -35.40) >DroYak_CAF1 9005 120 - 1 AUAGCUCCGGAAUCUCUGGAGAUAUAUCUCGUGGGAACCGAAUGAGUGCAGAUUCAAUGGAAUCGGAACUCAAUUCUGAUUUUAGUGGGGAGUUUUCAGAUAAUUGUGUAAGGAACUCGA (((.((((((.....)))))).)))..((((..(((((.((((((((...(((((....)))))...)))).)))).).))))..))))(((((((...(((....)))..))))))).. ( -32.40) >consensus AUAGCUCCGGAAUCUCUGGAGAUAUAUGUCGUGGAAGUCUGAUGAGUGCAGAUUCAAUGGAGUCCGAACUUAACUCAGAAUCUAGUGGGGAGCUUUUAGAUAAUAAUGAAUUGAUAGCGA (((.((((((.....)))))).))).((((.((((((((((((((((...(((((....)))))...)))))..))))..(((.....)))))))))))))).................. (-27.88 = -26.04 + -1.84)

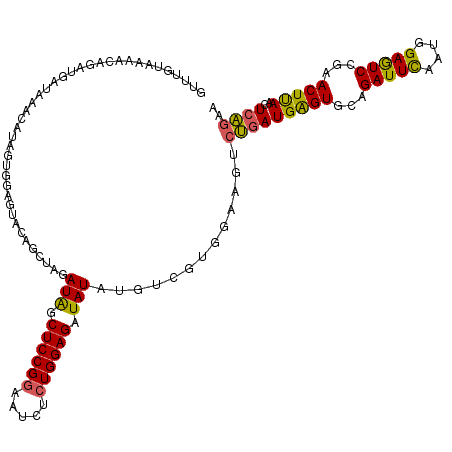
| Location | 2,390,290 – 2,390,407 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.92 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -21.00 |
| Energy contribution | -20.16 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2390290 117 - 22407834 AUUUGUGAUAA---GAAUAAGCCUAGUGGAGUACAGCUAGAUAGCUCCGGAAUAUCUGGAGGUAUAUGUCGUGGAAGUCUGAUGGGUGCAGAUUCAAUGGAGUCCGAACUUAACUCAGAA .((..(((((.---........(((((........)))))(((.((((((.....)))))).))).)))))..))..((((((((((.(.(((((....))))).).)))))..))))). ( -35.60) >DroSec_CAF1 8293 120 - 1 GUUUGUAAAGCAGGUGAUAAACAUAGUGGAGUACAGCUAGAUGGCUCCGGAAUCUCUGGAGAUAUAUGUCUUGGAAGUCUGAUGAGUGCAGAUUCAUUGGAGUCCGAACUUAACUCAGAA ((((((..........))))))......((((.......((((.((((((.....)))))).....))))(((((..((..((((((....))))))..)).))))).....)))).... ( -31.80) >DroSim_CAF1 8974 120 - 1 GUUUGUAAAACAGUUGAUAAACAUAAUGGAGUACAGGUAGAUAGCUCCGGAAUCUCUGGAGAUAUAUGUCUUGGAAGUCUGAUGAGUGCAGAUUUAUUGGAGUCCGAACUUAACUCACAA ((((((..........))))))....((((((..((((.((((.((((((.....)))))).....))))(((((..((..((((((....))))))..)).))))))))).)))).)). ( -32.60) >DroEre_CAF1 9207 108 - 1 CUUCAUAACACAGACGAUAAAC------------ACCUGGAUAGCUCCGGAAUCUAUGGAGGUAUAUGUCGUGGGAGCCGGAUGAGUGCAGAUUCAAUGGAAUCGGAACUCAAUUCUGAU ..........((((........------------...........(((((..(((((((.........)))))))..)))))(((((...(((((....)))))...)))))..)))).. ( -29.00) >DroYak_CAF1 9045 117 - 1 GC---UAAAAUAGAUGAUAAACAUGGUGGAGUAAACCUAGAUAGCUCCGGAAUCUCUGGAGAUAUAUCUCGUGGGAACCGAAUGAGUGCAGAUUCAAUGGAAUCGGAACUCAAUUCUGAU ..---.....((((.........((((........((.(((((.((((((.....))))))...)))))...))..))))..(((((...(((((....)))))...)))))..)))).. ( -30.90) >consensus GUUUGUAAAACAGAUGAUAAACAUAGUGGAGUACAGCUAGAUAGCUCCGGAAUCUCUGGAGAUAUAUGUCGUGGAAGUCUGAUGAGUGCAGAUUCAAUGGAGUCCGAACUUAACUCAGAA ........................................(((.((((((.....)))))).))).............(((((((((...(((((....)))))...)))))..)))).. (-21.00 = -20.16 + -0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:49 2006