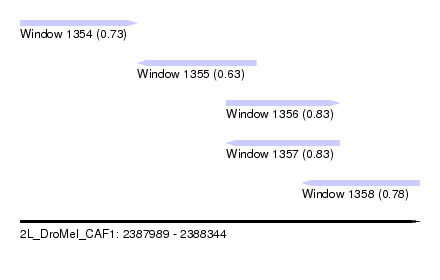
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,387,989 – 2,388,344 |
| Length | 355 |
| Max. P | 0.828937 |

| Location | 2,387,989 – 2,388,093 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.32 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -18.97 |
| Energy contribution | -21.00 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2387989 104 + 22407834 -CGCAUUGCCUAGUUGCAUUCU-CAACGAGUGCA--GCAAUUAGCAAGUAUUUUUGGAAGUGCCCCAAAGGAGGAGUCCUUUUUUGGG-CAAUCGCAUUAAGUGAUUCC -.(((((.....(((((((((.-....)))))))--))......((((....))))..)))))(((((((((((...)))))))))))-.((((((.....)))))).. ( -35.30) >DroSec_CAF1 5990 105 + 1 -CGCAUUGCCUAGUUGCAUUCU-CAACGAGUGCA--GCAAUUAGCAAGUAUUUUUGGAAGUGCCCCAAAGGAGGAGUCCUUUUUUGGGCCAAUCGCAUUAAGUGAUUCC -.(((((.....(((((((((.-....)))))))--))......((((....))))..)))))(((((((((((...)))))))))))..((((((.....)))))).. ( -35.30) >DroSim_CAF1 6665 105 + 1 -CGCAUUGCCUAGUUGCAUUCU-CAACGAGUGCA--GCAAUUAGCAAGUAUUUUUGGAAGUGCCCCAAAGGAGGAGUCCUUUUUUGGGCCAAUCGCAUUAAGUGAUUCC -.(((((.....(((((((((.-....)))))))--))......((((....))))..)))))(((((((((((...)))))))))))..((((((.....)))))).. ( -35.30) >DroEre_CAF1 6919 104 + 1 -CCCAUUGCCUAGUUGCAGUCC-CAGCGAUUGCA--GCAAUUAGCAAAUAUUUUUGAAAGUGCCCCAAAGUAUAACUCCUUUUUUGGG-CAAUCGCAUUAAGUGAUUCC -...(((((......)))))..-..((((((((.--(((.....((((....))))....))).((((((...........)))))))-)))))))............. ( -25.90) >DroYak_CAF1 6768 104 + 1 -CCCAUUGCCUAGUUGCAUUCU-CAACAAUUGCA--GCAAUUAGCAAAUAUUUUUGAAAGUGCCCCAAACGAGAACUCCUUUUUUGGG-CAAUCGCAUUAAGUGAUUCC -....((((.(((((((.....-...........--))))))))))).............(((((.(((.(((.....))).))))))-))(((((.....)))))... ( -22.99) >DroAna_CAF1 8085 89 + 1 ACUCAUUUUCAAGGUGCAUCUUCCAGCGAGGGCCGACCAAUUAGCAGCUA------------CCCCGAA-------CUCUUUUUUCGG-CAAUCGCAUUAAGUGAUUUC ..((((((....((........)).((((..(((((..((..((...(..------------....)..-------))..))..))))-)..))))...)))))).... ( -19.60) >consensus _CGCAUUGCCUAGUUGCAUUCU_CAACGAGUGCA__GCAAUUAGCAAGUAUUUUUGGAAGUGCCCCAAAGGAGGAGUCCUUUUUUGGG_CAAUCGCAUUAAGUGAUUCC ....(((((.....(((((((......)))))))..)))))......................((((((((((.....))))))))))..((((((.....)))))).. (-18.97 = -21.00 + 2.03)

| Location | 2,388,093 – 2,388,199 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -22.47 |
| Energy contribution | -21.23 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2388093 106 - 22407834 UCUGAGAUUUGCCAAGUGAAAAAAUA-----------ACAACAAGCGACUUUGCGAAUUAAUCUUCUUAAUGCCUGGGUCCCAUAACCAUGGGCACUGAGUGGAGUGCUUAGGACAC ((((((((((((.((((.........-----------..........)))).))))))...(((.((((.(((((((((......))).)))))).)))).)))...)))))).... ( -26.61) >DroPse_CAF1 12260 117 - 1 UCUGAGAUUUGCCGACUAAAAAACAACAUCACCCACAACAAAAUCAGAUUUCGAGAAUUAAUCUUCUUAAUGUUUGGGUCUCGUAGCCCCGAGUACUAAGUAGAAUGCUUAGACCUU ((((((((((....(((.............(((((.((((......(....)(((((......)))))..)))))))))((((......)))).....))).)))).)))))).... ( -21.30) >DroSim_CAF1 6770 106 - 1 UCUGAGAUUUGCCAAGUGAAAAAAUA-----------ACAACAAGCGACUUUGCGAAUUAAUCUUCUUAAUGCCCGGGUCCCAUAACCAUGGGCACUAAGUGGAGUGCUUAGGACAC ((((((((((((.((((.........-----------..........)))).))))))...(((.((((.(((((((((......))).)))))).)))).)))...)))))).... ( -29.41) >DroEre_CAF1 7023 106 - 1 UCUGAGAUUUGCCAAGUGAAAAAAAU-----------ACAACAAUCGACUUUGCGAAUUAAUCUUCUUAAUGCCUGGGCCCCAUAACCAUGGGCACUAAGUAGAGUGCUUAGGACAC ((((((((((((.((((((.......-----------.......)).)))).))))))...(((.((((.((((((((........)).)))))).)))).)))...)))))).... ( -25.64) >DroYak_CAF1 6872 106 - 1 UCUGAGAUUUGCCAAGUGAAAAAAAU-----------ACAACAAUCGACUUUGCGAAUUAAUCUUCUUAAUGCCUGGGCCCCAUAACCAUGGGCACUAAGUAGAGUGCUUAGGACAC ((((((((((((.((((((.......-----------.......)).)))).))))))...(((.((((.((((((((........)).)))))).)))).)))...)))))).... ( -25.64) >DroPer_CAF1 13508 117 - 1 UCUGAGAUUUGCCGACUAAAAAACAACAUCACCCACAACAAAAUCAGAUUUCGAGAAUUAAUCUUCUUAAUGUUUGGGCCUCGUAGCCCCGAGUACUAAGUAGAAUGCUUAGACCUU ((((((((((....(((..............((((.((((......(....)(((((......)))))..)))))))).((((......)))).....))).)))).)))))).... ( -20.30) >consensus UCUGAGAUUUGCCAAGUGAAAAAAAA___________ACAACAACCGACUUUGCGAAUUAAUCUUCUUAAUGCCUGGGCCCCAUAACCAUGGGCACUAAGUAGAGUGCUUAGGACAC ((((((((((((.((((..............................)))).))))))...(((.((((.(((((((((......))).)))))).)))).)))...)))))).... (-22.47 = -21.23 + -1.24)
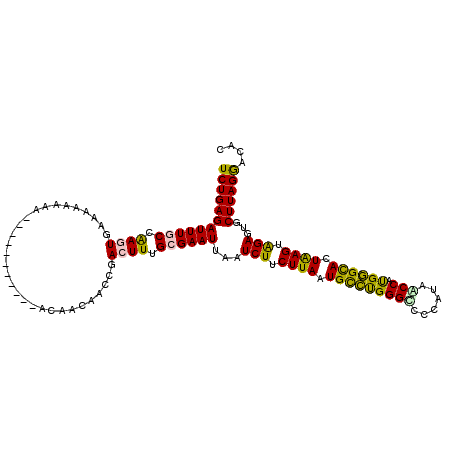
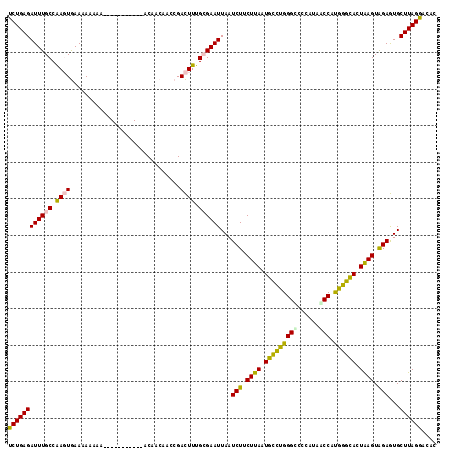
| Location | 2,388,172 – 2,388,273 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -23.64 |
| Consensus MFE | -12.72 |
| Energy contribution | -13.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2388172 101 + 22407834 U-----------UAUUUUUUCACUUGGCAAAUCUCAGAUAACGCUAUGCAUCAUUGUGUCAAAGAUUUGACAAAUCUUUGGG-AGGCCAACAAA---AAAAAAAGGAAAAAAUAAG (-----------(((((((((.((((((...(((((((....((...)).......((((((....))))))....))))))-).)))).....---......)))))))))))). ( -23.90) >DroSec_CAF1 6174 104 + 1 U-----------UAUUUUUUCACUUGGCAAAUCUCAGAUAACGCUAUGCAUCAUUGUGUCAAAGAUUUGACAAAUCUUUGGG-AGGCCAACCAAAAAAAAAAAAGGAAAAAAUAAG (-----------(((((((((.((((((...(((((((....((...)).......((((((....))))))....))))))-).))))..............)))))))))))). ( -23.63) >DroSim_CAF1 6849 104 + 1 U-----------UAUUUUUUCACUUGGCAAAUCUCAGAUAACGCUAUGCAUCAUUGUGUCAAAGAUUUGACAAAUCUUUGGG-AGGCCAACAAAAAAAAAAAAAGGAAAAAAUAAG (-----------(((((((((.((((((...(((((((....((...)).......((((((....))))))....))))))-).))))..............)))))))))))). ( -23.63) >DroEre_CAF1 7102 90 + 1 U-----------AUUUUUUUCACUUGGCAAAUCUCAGAUAACGCUAUGCAUCAUUGUGUCAAAGAUUUGACAAAUCCUUGGG-AGGCCGACAAAA--------------AAAUAAG (-----------((((((((...(((((...((((((.....((...)).......((((((....)))))).....)))))-).))))).))))--------------))))).. ( -21.30) >DroYak_CAF1 6951 91 + 1 U-----------AUUUUUUUCACUUGGCAAAUCUCAGAUAACGCCAUGCAUCAUUGUGUCAAAGAUUUGACAAAUCUUUGCG-AGGCCAACAAAA-------------AAAAUAAA (-----------((((((((...(((((....(((.....((((.(((...))).))))((((((((.....)))))))).)-)))))))..)))-------------)))))).. ( -20.40) >DroPer_CAF1 13587 115 + 1 UUGUGGGUGAUGUUGUUUUUUAGUCGGCAAAUCUCAGAUAACGUUAUGCAUCAUUGUGUCAAAGAAUUGACAAAUCUUUGGCCAUGCGAGCA-GGGCGGAAGAAGGAAAAAAUAAG ..(((.((((((((((((...((.........)).)))))))))))).))).....((((((....))))))..(((((.(((.((....))-.))).)))))............. ( -29.00) >consensus U___________UAUUUUUUCACUUGGCAAAUCUCAGAUAACGCUAUGCAUCAUUGUGUCAAAGAUUUGACAAAUCUUUGGG_AGGCCAACAAAA__AAAAAAAGGAAAAAAUAAG .......................(((((............((((.(((...))).))))((((((((.....)))))))).....))))).......................... (-12.72 = -13.00 + 0.28)

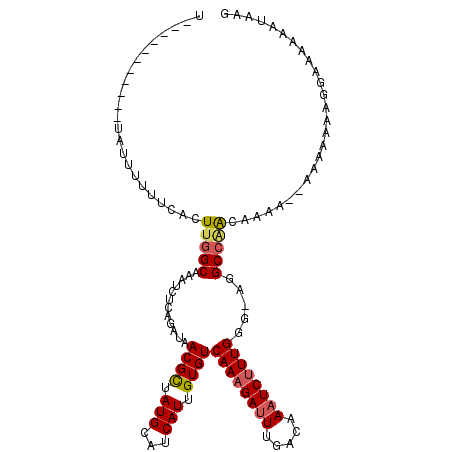
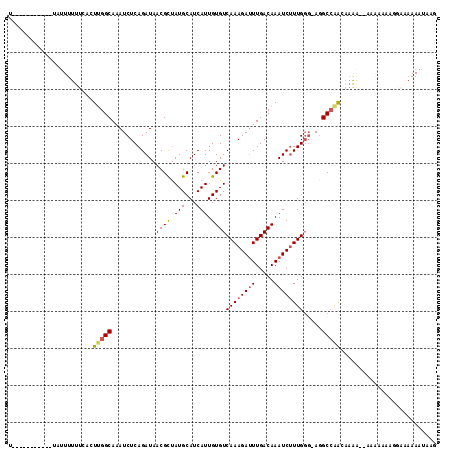
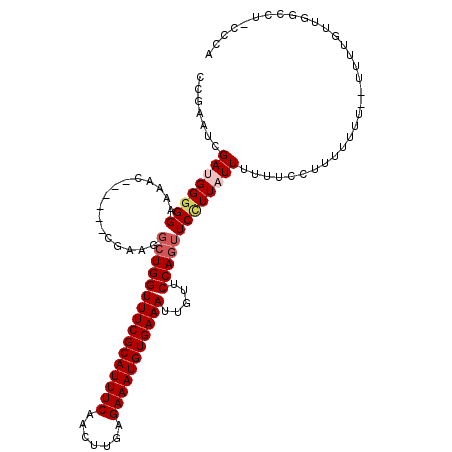
| Location | 2,388,172 – 2,388,273 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -14.40 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2388172 101 - 22407834 CUUAUUUUUUCCUUUUUUU---UUUGUUGGCCU-CCCAAAGAUUUGUCAAAUCUUUGACACAAUGAUGCAUAGCGUUAUCUGAGAUUUGCCAAGUGAAAAAAUA-----------A ..(((((((((........---....((((...-.))))..(((((.((((((((.((.....((((((...)))))))).)))))))).))))))))))))))-----------. ( -23.00) >DroSec_CAF1 6174 104 - 1 CUUAUUUUUUCCUUUUUUUUUUUUGGUUGGCCU-CCCAAAGAUUUGUCAAAUCUUUGACACAAUGAUGCAUAGCGUUAUCUGAGAUUUGCCAAGUGAAAAAAUA-----------A ..(((((((((.........((((((.......-.))))))(((((.((((((((.((.....((((((...)))))))).)))))))).))))))))))))))-----------. ( -24.80) >DroSim_CAF1 6849 104 - 1 CUUAUUUUUUCCUUUUUUUUUUUUUGUUGGCCU-CCCAAAGAUUUGUCAAAUCUUUGACACAAUGAUGCAUAGCGUUAUCUGAGAUUUGCCAAGUGAAAAAAUA-----------A ..(((((((((...............((((...-.))))..(((((.((((((((.((.....((((((...)))))))).)))))))).))))))))))))))-----------. ( -23.00) >DroEre_CAF1 7102 90 - 1 CUUAUUU--------------UUUUGUCGGCCU-CCCAAGGAUUUGUCAAAUCUUUGACACAAUGAUGCAUAGCGUUAUCUGAGAUUUGCCAAGUGAAAAAAAU-----------A ..(((((--------------((((.....(((-....)))(((((.((((((((.((.....((((((...)))))))).)))))))).))))).))))))))-----------) ( -19.90) >DroYak_CAF1 6951 91 - 1 UUUAUUUU-------------UUUUGUUGGCCU-CGCAAAGAUUUGUCAAAUCUUUGACACAAUGAUGCAUGGCGUUAUCUGAGAUUUGCCAAGUGAAAAAAAU-----------A ..((((((-------------((((((((((((-((((((((((.....)))))))).)...(((((((...)))))))..)))....))))).))))))))))-----------) ( -24.30) >DroPer_CAF1 13587 115 - 1 CUUAUUUUUUCCUUCUUCCGCCC-UGCUCGCAUGGCCAAAGAUUUGUCAAUUCUUUGACACAAUGAUGCAUAACGUUAUCUGAGAUUUGCCGACUAAAAAACAACAUCACCCACAA .............((((..(((.-((....)).)))..))))..((((((....))))))...(((((.......(((..((........))..))).......)))))....... ( -16.54) >consensus CUUAUUUUUUCCUUUUUUU__UUUUGUUGGCCU_CCCAAAGAUUUGUCAAAUCUUUGACACAAUGAUGCAUAGCGUUAUCUGAGAUUUGCCAAGUGAAAAAAAA___________A ..........................(((((.....((((((((.....))))))))...(((((((((...))))))).))......)))))....................... (-14.40 = -14.98 + 0.58)
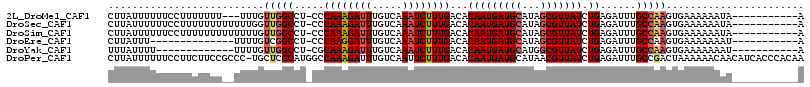
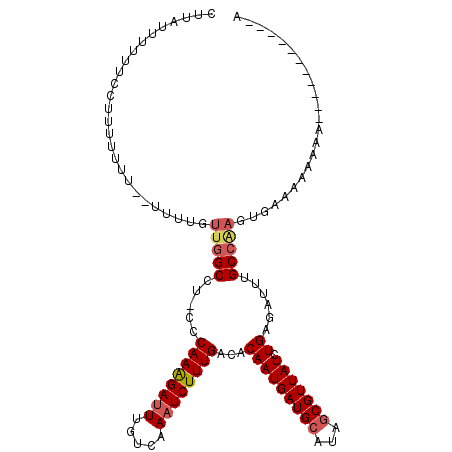
| Location | 2,388,239 – 2,388,344 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.99 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -17.33 |
| Energy contribution | -18.06 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2388239 105 - 22407834 CCGAAUCGAUGGGGGAAAAC------CGAAGGCUGGUUUCGCAUUUCAACUUGAGAAAUGUGAAAUUGUUCCAGUUCCUUAUUUUUUCCUUUUUUU---UUUGUUGGCCU-CCCA ((((......(((((((((.------..(((((((((((((((((((.......))))))))))).....))))..))))...)))))))))....---....))))...-.... ( -29.74) >DroPse_CAF1 12417 104 - 1 -ACAAUCGAGGG-GGAAA-C------C-AGGCCUGGUUUCGCAUUUCAACUUGAGAAAUGUGAAAUUGUUCCAAUUCCUUAUUUUUUCCUUCUUCCGCCC-UGCUCGCAUGGCCA -......(((((-..(((-.------.-(((..((((((((((((((.......))))))))))).....)))...)))...)))..)))))....(((.-((....)).))).. ( -31.50) >DroSec_CAF1 6241 108 - 1 CCGAAUCGAAGGGGGAAAAC------CGAAGGCUGGUUUCGCAUUUCAACUUGAGAAAUGUGAAAUUGUUCCAGUUCCUUAUUUUUUCCUUUUUUUUUUUUGGUUGGCCU-CCCA (((((..((((((((((((.------..(((((((((((((((((((.......))))))))))).....))))..))))...))))))))))))...))))).(((...-.))) ( -34.30) >DroSim_CAF1 6916 108 - 1 CCGAAUCGAUGGGGGAAAAC------CGAAGGCUGGUUUCGCAUUUCAACUUGAGAAAUGUGAAAUUGUUCCAGUUCCUUAUUUUUUCCUUUUUUUUUUUUUGUUGGCCU-CCCA .((((..((.(((((((((.------..(((((((((((((((((((.......))))))))))).....))))..))))...)))))))))...))..)))).(((...-.))) ( -28.40) >DroEre_CAF1 7169 94 - 1 CCAAAUCGAUGGGGGAAAUC------CGACGGCUGGUUUCGCAUUUCAACUUGAGAAAUGUGAAAUUGUUCCAGUUCCUUAUUU--------------UUUUGUCGGCCU-CCCA (((......)))((((...(------(((((((((((((((((((((.......))))))))))).....))))).........--------------...))))))..)-))). ( -31.50) >DroYak_CAF1 7018 101 - 1 UCAAAUCGAUGGGGGAAACCGAAAAGCGACGGCUGGUUUCGCAUUUCAACUUGAGAAAUGUGAAAUUGUUCCAGUUCUUUAUUUU-------------UUUUGUUGGCCU-CGCA ............((....)).....((((.(((((((((((((((((.......)))))))))))))....(((...........-------------..)))..)))))-))). ( -29.12) >consensus CCGAAUCGAUGGGGGAAAAC______CGAAGGCUGGUUUCGCAUUUCAACUUGAGAAAUGUGAAAUUGUUCCAGUUCCUUAUUUUUUCCUUUUUUU__UUUUGUUGGCCU_CCCA .......((((((((................((((((((((((((((.......))))))))))).....)))))))))))))................................ (-17.33 = -18.06 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:45 2006