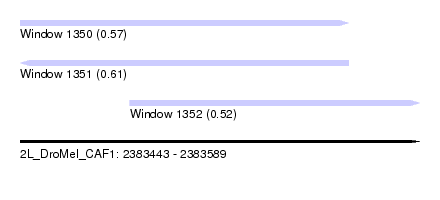
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,383,443 – 2,383,589 |
| Length | 146 |
| Max. P | 0.605280 |

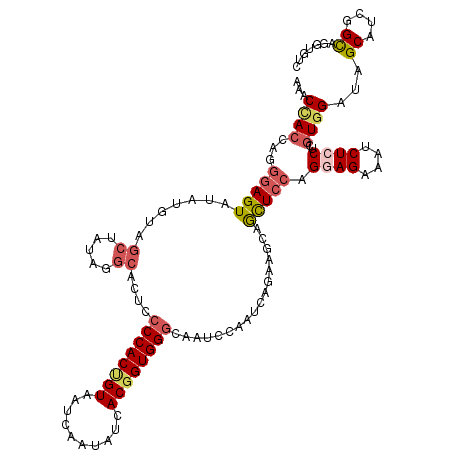
| Location | 2,383,443 – 2,383,563 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.67 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -25.59 |
| Energy contribution | -24.85 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
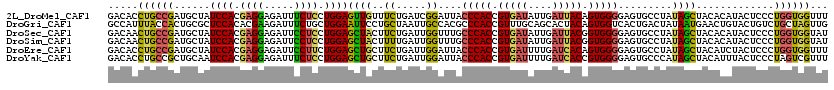
>2L_DroMel_CAF1 2383443 120 + 22407834 GACACCUGCCGAUGCUAUCCACGAGGAGAUUUCUCCUGGAGUUGUUUCUGAUCGGAUUACCCACCGUGAUAUUGAUUACAGUGGGGAGUGCCUAUAGCUACACAUACUCCCUGGUGGUUU ..((((.................(((((....)))))((((((((..(((...(((((.(((((.(((((....))))).))))).))).))..)))..)))...)))))..)))).... ( -36.80) >DroGri_CAF1 2986 120 + 1 GCCAUUUACCACUGCGCUCCACACGAAGAUUUCUGCUGGAAUUCCUGCUAAUUGCCACGCCCACCGUUUGCAGCACUACAGUGGUCACUGACUAUAAUGAACUGUACUGUCUGCUAGUUG ..((.(.(((((((.(((.((.(((.((....))((.((....)).))................))).)).)))....))))))).).)).........(((((((.....))).)))). ( -22.30) >DroSec_CAF1 1441 120 + 1 GACAACUGCCGAUGCUAUCCACGAGGAGAUUCCUCCUGGAGCUACUUCUGAUUGGUUUGCCCACCGUGAUAUUGAUUACGGUGGGGAGUGCCUAUAGCUACACAUACUCCCUGGUGGUAU ....((..(((......((((.((((.....)))).))))(((((....)...(((((.(((((((((((....))))))))))).)).)))..)))).............)))..)).. ( -43.60) >DroSim_CAF1 2110 120 + 1 GACAACUGCCGAUGCUAUCCACGAGGAGAUUCCUCCUGGAGCUACUUUUGAUUGGUUUGCCCACCGUGAUAUUGAUUACGGUGGGGAGUGCCUAUAGCUACACAUACUCCCUGGUGGUAU ....((..(((......((((.((((.....)))).))))(((((....)...(((((.(((((((((((....))))))))))).)).)))..)))).............)))..)).. ( -43.60) >DroEre_CAF1 2329 120 + 1 GACACCUGCCGAUGCUAUCCACGAGGAGAUUCCUUCUGGAGCUGCUUCUGAUUGGAUUACCCACCGUGAUUUUGAUCACAGUGGGGAGUGCCUAUAGCUACAUCUACUCCCUGGUGGUUU ....((..(.((.((..((((.((((.....)))).))))...)).)).)...)).....((((((((((....)))))...((((((((..............)))))))))))))... ( -40.54) >DroYak_CAF1 2133 120 + 1 GACACCUGCCGCUGCAAUCCACGAGGAGAUUUCUCCUGGAGCUGCUUCUGAUUGGAUUACCCACCGUGAUUUUGAUCACCGUGGGGAGUGCCCAUAGCUACAUUUACUCCCUAGUCGUUU (((..........(((.((((...((((....))))))))..)))....((((((....(((((.(((((....))))).)))))(((((..............))))).))))))))). ( -36.34) >consensus GACACCUGCCGAUGCUAUCCACGAGGAGAUUCCUCCUGGAGCUGCUUCUGAUUGGAUUACCCACCGUGAUAUUGAUUACAGUGGGGAGUGCCUAUAGCUACACAUACUCCCUGGUGGUUU .....((((((......((((.((((.....)))).))))((((..((.....))....(((((.(((((....))))).))))).........)))).............))))))... (-25.59 = -24.85 + -0.74)
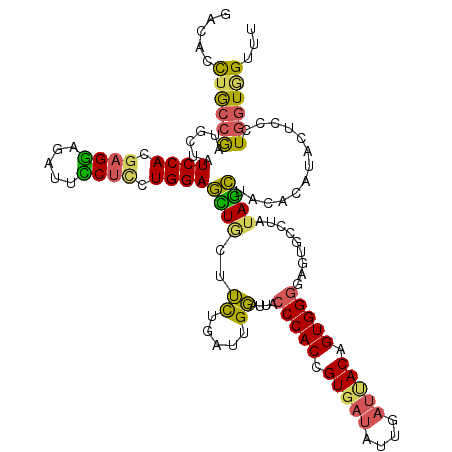
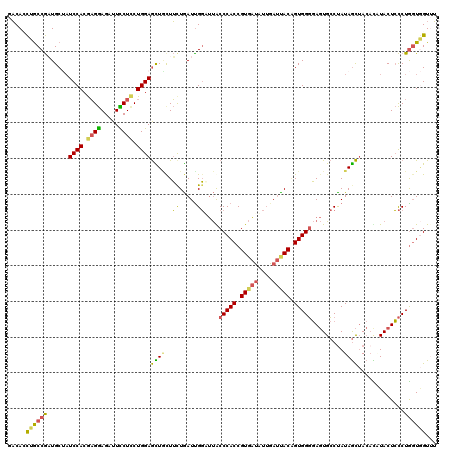
| Location | 2,383,443 – 2,383,563 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.67 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -24.34 |
| Energy contribution | -25.23 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2383443 120 - 22407834 AAACCACCAGGGAGUAUGUGUAGCUAUAGGCACUCCCCACUGUAAUCAAUAUCACGGUGGGUAAUCCGAUCAGAAACAACUCCAGGAGAAAUCUCCUCGUGGAUAGCAUCGGCAGGUGUC ...((((...(((((.(((....((...((.....((((((((..........))))))))....))....))..))))))))(((((....))))).))))...(((((....))))). ( -38.20) >DroGri_CAF1 2986 120 - 1 CAACUAGCAGACAGUACAGUUCAUUAUAGUCAGUGACCACUGUAGUGCUGCAAACGGUGGGCGUGGCAAUUAGCAGGAAUUCCAGCAGAAAUCUUCGUGUGGAGCGCAGUGGUAAAUGGC ......((((.((.(((((((((((......)))))..)))))).))))))...((.((.((.(.(((....(.((((.(((.....))).))))).))).).)).)).))......... ( -32.70) >DroSec_CAF1 1441 120 - 1 AUACCACCAGGGAGUAUGUGUAGCUAUAGGCACUCCCCACCGUAAUCAAUAUCACGGUGGGCAAACCAAUCAGAAGUAGCUCCAGGAGGAAUCUCCUCGUGGAUAGCAUCGGCAGUUGUC ...((((...(((((..((((........))))..((((((((..........)))))))).................)))))..((((.....))))))))...((....))....... ( -37.00) >DroSim_CAF1 2110 120 - 1 AUACCACCAGGGAGUAUGUGUAGCUAUAGGCACUCCCCACCGUAAUCAAUAUCACGGUGGGCAAACCAAUCAAAAGUAGCUCCAGGAGGAAUCUCCUCGUGGAUAGCAUCGGCAGUUGUC ...((((...(((((..((((........))))..((((((((..........)))))))).................)))))..((((.....))))))))...((....))....... ( -37.00) >DroEre_CAF1 2329 120 - 1 AAACCACCAGGGAGUAGAUGUAGCUAUAGGCACUCCCCACUGUGAUCAAAAUCACGGUGGGUAAUCCAAUCAGAAGCAGCUCCAGAAGGAAUCUCCUCGUGGAUAGCAUCGGCAGGUGUC ...((((...(((((...(((..((((.((.....(((((((((((....)))))))))))....)).)).))..)))))))).((.((.....))))))))...(((((....))))). ( -40.60) >DroYak_CAF1 2133 120 - 1 AAACGACUAGGGAGUAAAUGUAGCUAUGGGCACUCCCCACGGUGAUCAAAAUCACGGUGGGUAAUCCAAUCAGAAGCAGCUCCAGGAGAAAUCUCCUCGUGGAUUGCAGCGGCAGGUGUC ..............((..(((.(((.((((.....(((((.(((((....))))).)))))...)))).......((((.((((.(((.......))).))))))))))).)))..)).. ( -38.60) >consensus AAACCACCAGGGAGUAUAUGUAGCUAUAGGCACUCCCCACUGUAAUCAAUAUCACGGUGGGCAAUCCAAUCAGAAGCAGCUCCAGGAGAAAUCUCCUCGUGGAUAGCAUCGGCAGGUGUC ...((((...(((((.......((.....))....((((((((..........)))))))).................))))).((((....))))..))))...((....))....... (-24.34 = -25.23 + 0.89)

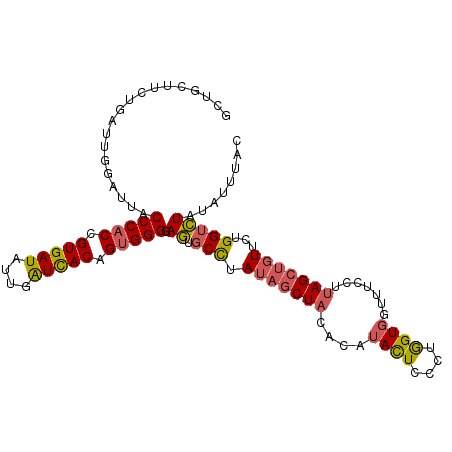
| Location | 2,383,483 – 2,383,589 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2383483 106 + 22407834 GUUGUUUCUGAUCGGAUUACCCACCGUGAUAUUGAUUACAGUGGGGAGUGCCUAUAGCUACACAUACUCCCUGGUGGUUUCCUUAGCUGUUCUGGUCUAUAUUUAU .........(((((((...(((((.(((((....))))).))))).(((......((((((.((.......))))))))......)))..)))))))......... ( -28.30) >DroSec_CAF1 1481 106 + 1 GCUACUUCUGAUUGGUUUGCCCACCGUGAUAUUGAUUACGGUGGGGAGUGCCUAUAGCUACACAUACUCCCUGGUGGUAUCCUUAGCUGUUCUGGUCUAUAUUUAC .........(((((((((.(((((((((((....))))))))))).)).))).(((((((...((((((....).)))))...)))))))...))))......... ( -33.20) >DroSim_CAF1 2150 106 + 1 GCUACUUUUGAUUGGUUUGCCCACCGUGAUAUUGAUUACGGUGGGGAGUGCCUAUAGCUACACAUACUCCCUGGUGGUAUCCUUAGCUGUUCUGGUCUAUAUUUAC .........(((((((((.(((((((((((....))))))))))).)).))).(((((((...((((((....).)))))...)))))))...))))......... ( -33.20) >DroEre_CAF1 2369 106 + 1 GCUGCUUCUGAUUGGAUUACCCACCGUGAUUUUGAUCACAGUGGGGAGUGCCUAUAGCUACAUCUACUCCCUGGUGGUUUUAUUAGCUGUUCUGGUCUAUAUUUAC .........(((..((...(((((.(((((....))))).)))))........(((((((...(((((....)))))......)))))))))..)))......... ( -30.60) >DroYak_CAF1 2173 106 + 1 GCUGCUUCUGAUUGGAUUACCCACCGUGAUUUUGAUCACCGUGGGGAGUGCCCAUAGCUACAUUUACUCCCUAGUCGUUUUCUUAGGUGUUGUGGUCUAUAUUUAC ............((((((((.(((((((((....)))))..(((((((((..............)))))))))............))))..))))))))....... ( -28.64) >DroAna_CAF1 2470 106 + 1 GUUCCUGUUGAUUGGAUUGCCCUCUGUGAUUUUGGUCACCGUGGGAAAUGCUUACAGCUCCACCUAUUCUUGGGUGGGCUCCAUAGCCAUCAUGGGUUAUCUAUAC ............(((((.((((...(((((....))))).((((.....(((...(((.(((((((....))))))))))....)))..)))))))).)))))... ( -33.60) >consensus GCUGCUUCUGAUUGGAUUACCCACCGUGAUAUUGAUCACAGUGGGGAGUGCCUAUAGCUACACAUACUCCCUGGUGGUUUCCUUAGCUGUUCUGGUCUAUAUUUAC ...................(((((.(((((....))))).))))).((.(((.(((((((....((((....)))).......)))))))...)))))........ (-19.50 = -19.58 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:39 2006