
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,382,489 – 2,382,598 |
| Length | 109 |
| Max. P | 0.716745 |

| Location | 2,382,489 – 2,382,598 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -20.48 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2382489 109 - 22407834 AAACUGAAGCCCAAGGAGUGUAU-UUAAACAUUUCUUACCUUUCAUAGGAAACACGCUGUACAUCGUACUUAUCGUUGGCCUCCAGGGGGGCCAGCAGCUCGUCUUCAUU ....((((((..(((((((((..-....))))))))).(((.....)))......(((((((...)))).....(((((((((....))))))))))))..).))))).. ( -35.50) >DroVir_CAF1 1877 108 - 1 UAACUACAGCUUAAGGAAUAUUUAAUUA-A-AAAAUUACCGUUCGUAUGAGACACGCUGCACAACGUACUCCUCAUUGGCCUCCAACUGGGCUAGCAGAUCGUUCUCGUU ...............((((...(((((.-.-..)))))..))))..((((((.(((((((.....(.......)..((((((......))))))))))..))))))))). ( -19.70) >DroSec_CAF1 487 109 - 1 AAACGGAAGCCCAAGGAGCGUAU-UUAAACAUUUUUUACCUUUCAUAGGAAACGCGCUGUACAUCGUACUUAUCGUUGGCCUCCAGGGGGGCCAGCAGCUCGUCUUCAUU ....(((((...((((((.((..-....)).))))))..)))))...(((.(((.(((((((...)))).....(((((((((....)))))))))))).))).)))... ( -33.20) >DroSim_CAF1 1157 109 - 1 AAACGGAAGCCCAAGGAGCAUAU-UUAAACAUUUUUUACCUUUCAUAGGAAACGCGCUGUACAUCGUACUUAUCGUUGGCCUCCAGGGGGGCCAGCAGCUCGUCUUCAUU ....((....))..((((.....-..............(((.....)))..(((.(((((((...)))).....(((((((((....)))))))))))).)))))))... ( -31.30) >DroEre_CAF1 1375 109 - 1 AAACUAAAGACUAGGGAGAGGUU-UUGAUCAUUCCUUACCUCUCGUAGGACACACGCUGCACAGCGUACUUAUCGUUGGCCUCCAGGGGGGCCAGCAGCUCGUCUUCAUU ......(((((.((.(((((((.-..((....))...)))))))(((((....(((((....))))).))))).(((((((((....)))))))))..)).))))).... ( -42.20) >DroYak_CAF1 1178 109 - 1 AAACUGAAGACUAAGUAGCGGCU-UUUAUCAUUCCUUACCUCUCAUAGGACACACGCUGCACGGCGUACUUAUCGUUGGCCUCCAGGGGGGCCAGCAGCUCGUCUUCAUU ....((((((((((((.......-........((((..........))))...(((((....))))))))))..(((((((((....))))))))).....))))))).. ( -38.30) >consensus AAACUGAAGCCCAAGGAGCGUAU_UUAAACAUUUCUUACCUUUCAUAGGAAACACGCUGCACAUCGUACUUAUCGUUGGCCUCCAGGGGGGCCAGCAGCUCGUCUUCAUU ...............((((...................(((.....)))....(((........))).......(((((((((....))))))))).))))......... (-20.48 = -21.12 + 0.64)

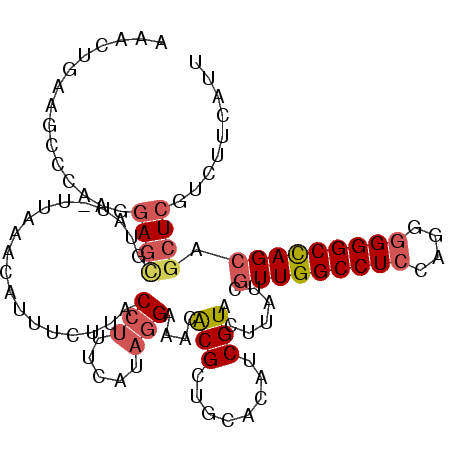
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:35 2006