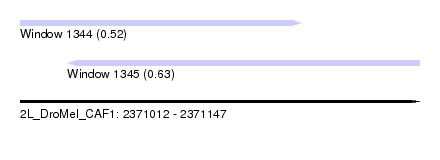
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,371,012 – 2,371,147 |
| Length | 135 |
| Max. P | 0.631431 |

| Location | 2,371,012 – 2,371,107 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.73 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.53 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
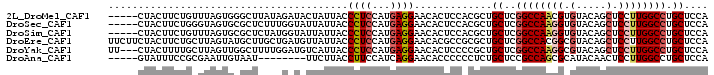
>2L_DroMel_CAF1 2371012 95 + 22407834 -----CUACUUCUGUUUAGUGGGCUUAUAGAUACUAUUACCCUCCAUGAGGAACACUCCACGCUGCUCGGCCAACGUGUACAGCUCCUUGGCCUGCUCCA -----............((..((((...((..........((((...))))..........((((..((.....))....))))..)).))))..))... ( -20.80) >DroSec_CAF1 77827 95 + 1 -----CUACUUCUGGGUAGUGCGCUCUUUGGUAUUAUUACCCUCCAUGAGGAACACUCCACGCUGCUCGGCCAAGGUGUACAGCUCCUUGGCCUGCUCCA -----........((((((((........((((....))))........(((....))).))))))))((((((((.(.....).))))))))....... ( -33.50) >DroSim_CAF1 77253 95 + 1 -----CUACUUCUGUUUAGUGCGCUCUAUGGUAUUAUUACCCUCCAUGAGGAACACUCCACGCUGCUCGGCCAAGGUGUACAGCUCCUUGGCCUGCUCCA -----........((..((((..(..(((((............)))))..)..))))..))(..((..((((((((.(.....).)))))))).))..). ( -29.70) >DroEre_CAF1 83161 100 + 1 UUCUUCUACUUCUGCUUAGUAUGCUUGCUGAUGUUAUUACCCUCCAUGAGGAACACGCCGCGCUGCUCGGCCACGGCGUACAGCUCCUUGGCCUGCUCCA .................((((.(((.((((..........((((...))))...((((((.(((....)))..)))))).)))).....))).))))... ( -27.70) >DroYak_CAF1 81908 97 + 1 UU---CUACUUUUGCUUAGUUGGCUUUUGGAUGUCAUUACCCUCCAUGAGGAACACUCCCCGCUGCUCGGCCAAGGCGUACAGCUCCUUGGCCUGCUCCA ..---........(((.....)))...((((.........((((...)))).............((..((((((((.(.....).)))))))).)))))) ( -26.40) >DroAna_CAF1 85050 87 + 1 -----GUAUUUCCGCGAAUUGUAAU--------UUCUUACCUUCCAUCAGGAACACCCCCCUCUGCUCCGCCAGCGCAUACAACUCCUUGGCCUGCUCCA -----(((...(((.((.(((((..--------........((((....))))...........(((.....)))...))))).))..)))..))).... ( -10.90) >consensus _____CUACUUCUGCUUAGUGCGCUUUUUGAUAUUAUUACCCUCCAUGAGGAACACUCCACGCUGCUCGGCCAAGGCGUACAGCUCCUUGGCCUGCUCCA ........................................((((...)))).............((..((((((((.(.....).)))))))).)).... (-15.78 = -16.53 + 0.75)

| Location | 2,371,028 – 2,371,147 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.39 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -24.99 |
| Energy contribution | -24.18 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
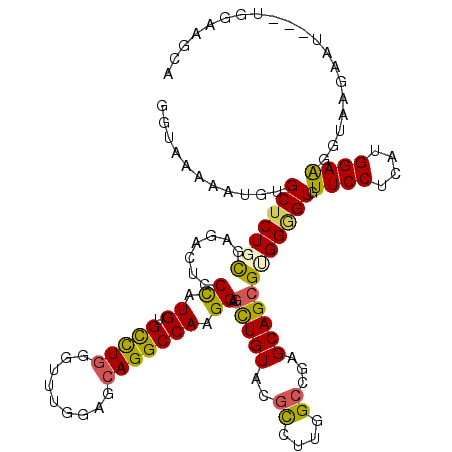
>2L_DroMel_CAF1 2371028 119 - 22407834 GGUAAAAACGUGCUCUGUGAGACUCCCAUGUGCUUGAGCGUGGAGCAGGCCAAGGAGCUGUACACGUUGGCCGAGCAGCGUGGAGUGUUCCUCAUGGAGGGUAAUAGUAUCUAUAAGCC ((((......(.((((((((((((((.(((((((((((((((..((((.(....)..)))).))))))...)))))).))))))))....))))))))).)......))))........ ( -40.90) >DroPse_CAF1 139008 116 - 1 GGAAAGCAUGUGCUCUGCGAGACACCUAUGUGUCUGGGUUUGGAACAGGCCAAGGAGCUGUGUGUCUUGGCAGAGCAGCGUGGGGUUUUCCUGAUGGAAGGUAAGACU---UGGGAGCG ......((((((((((((((((((((...((..((.((((((...)))))).))..)).).))))))).)))))))).))))((((((((((......))).))))))---)....... ( -45.40) >DroEre_CAF1 83182 119 - 1 GGUAAAAAUGUGCUCUGUGAGACUCCCAUGUGCUUGGGCUUGGAGCAGGCCAAGGAGCUGUACGCCGUGGCCGAGCAGCGCGGCGUGUUCCUCAUGGAGGGUAAUAACAUCAGCAAGCA ..........((((.(((...(((((((((......((((((...)))))).((((((...((((((((.(......)))))))))))))))))))).))))....)))..)))).... ( -44.50) >DroYak_CAF1 81926 119 - 1 GGUAAAAAUGUGCUCUGUGAGACUCCCAUGUGCUUGGGUUUGGAGCAGGCCAAGGAGCUGUACGCCUUGGCCGAGCAGCGGGGAGUGUUCCUCAUGGAGGGUAAUGACAUCCAAAAGCC (((.........(((((((((((((((...(((((..((.....)).((((((((.(.....).)))))))))))))...))))))....)))))))))((.........))....))) ( -45.10) >DroAna_CAF1 85066 111 - 1 GGGAAGCAUGUGCUCUGCGAGACGCCCAUGUGCCUAAGUCUGGAGCAGGCCAAGGAGUUGUAUGCGCUGGCGGAGCAGAGGGGGGUGUUCCUGAUGGAAGGUAAGAA--------AUUA (((((((.(.((((((((.((.(((.((....(((..(((((...)))))..)))...))...))))).)))))))).).....)).)))))...............--------.... ( -35.30) >DroPer_CAF1 149901 116 - 1 GGAAAGCAUGUGCUCUGCGAGACACCUAUGUGUCUGGGUUUGGAACAGGCCAAGGAGCUGUGUGUCUUGGCAGAGCAGCGUGGGGUUUUCCUGAUGGAAGGUAAGACU---UGGGAGCG ......((((((((((((((((((((...((..((.((((((...)))))).))..)).).))))))).)))))))).))))((((((((((......))).))))))---)....... ( -45.40) >consensus GGUAAAAAUGUGCUCUGCGAGACUCCCAUGUGCCUGGGUUUGGAGCAGGCCAAGGAGCUGUACGCCUUGGCCGAGCAGCGUGGGGUGUUCCUCAUGGAAGGUAAGAAU___UGGAAGCA ...........(((((((.......((.((.(((((.........))))))).)).(((((..((....))...)))))))))))).((((....)))).................... (-24.99 = -24.18 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:32 2006