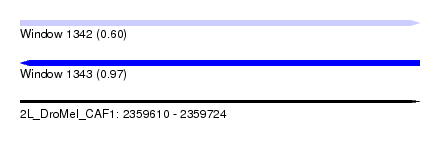
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,359,610 – 2,359,724 |
| Length | 114 |
| Max. P | 0.972406 |

| Location | 2,359,610 – 2,359,724 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -21.72 |
| Energy contribution | -23.58 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2359610 114 + 22407834 UAUAAAUUGCAACUAUCACACGUGUCC-GCCGCGACCCGGAGUCGAAUCAUCCGCGUCCGUAUCCGGAUCUGGAGUCCGAGCUCCGGAGCCGUAGA--GACACACACAUUGCCACAA .....................(((((.-.(..((.((((((((((.....((((.(((((....))))).))))...))).)))))).).))..).--))))).............. ( -36.80) >DroPse_CAF1 124734 99 + 1 UAUAAAUUGCAACUAUCACACGUGUCC-GUCGCAGCUUGGAGGCGGGG----------GGUAGGCGAUUC-----UCCGCACUCCAGAGCCGGACA--GACACACACAUUGCCGCAG .......(((...........(((((.-(((.(.((((((((((((((----------(((.....))))-----))))).))))).))).)))).--))))).(.....)..))). ( -37.80) >DroSim_CAF1 65642 114 + 1 UAUAAAUUGCAACUAUCACACGUGUCC-GCCGCGACCCGGAGUCGAAUCAUCCGCGUCCGUAAUCGGAUCUGGAGUCCGGGCUCCGGAGCCGCAGA--GACACACACAUUGCCACAA .....................((((((-((.((...((((((((......((((.(((((....))))).)))).....)))))))).)).)).).--))))).............. ( -39.40) >DroEre_CAF1 71474 114 + 1 UAUAAAUUGCAACUAUCACACGUGUCC-GCCGCGGCCCGGAGUCGAAUCAUCCGCGUCCGUAGCCGGAUCUGGAGUCCGGGCUCCGGAGCCGGAGA--GACACACACAUUGCCACAA .....................((((((-..(.((((((((((((......((((.(((((....))))).)))).....)))))))).))))).).--))))).............. ( -43.00) >DroYak_CAF1 70275 114 + 1 UAUAAAUUGCAACUAUCACACGUGUCC-GCCGCGACCCGGAGUCGAAUCAUCCGCGUCCAUAUCCGGAUCUGGAGUCCGGCCUCCGGAGCCGGAGA--GACACACACAUUGCCACAA .....................((((((-.(((.(..(((((((((.....((((.((((......)))).))))...))).))))))..)))).).--))))).............. ( -37.70) >DroAna_CAF1 73580 111 + 1 UAUAAAUUGCAACUAUCACACGUGUCCCAUCCCGACUUUAAGUCGAAUCCGUAGACUCCAA----UUAUCCGUAGACCGA--CCCGGAGCCGGAGAGAGACACACUCAUUGCCACAU .....................(((((.(....((((.....))))..........((((..----...((((........--..))))...)))).).))))).............. ( -21.50) >consensus UAUAAAUUGCAACUAUCACACGUGUCC_GCCGCGACCCGGAGUCGAAUCAUCCGCGUCCGUAGCCGGAUCUGGAGUCCGGGCUCCGGAGCCGGAGA__GACACACACAUUGCCACAA .....................(((((......((.((((((((((.....((((.(((((....))))).))))...))).)))))).).))......))))).............. (-21.72 = -23.58 + 1.86)

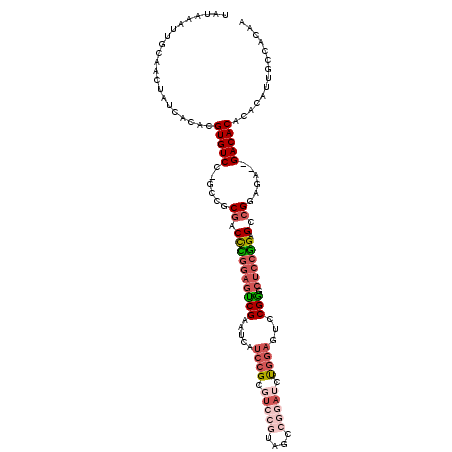
| Location | 2,359,610 – 2,359,724 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -48.78 |
| Consensus MFE | -29.45 |
| Energy contribution | -31.33 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2359610 114 - 22407834 UUGUGGCAAUGUGUGUGUC--UCUACGGCUCCGGAGCUCGGACUCCAGAUCCGGAUACGGACGCGGAUGAUUCGACUCCGGGUCGCGGC-GGACACGUGUGAUAGUUGCAAUUUAUA (((..((.(((..((((((--((..((((.((((((.(((((((((.(.((((....)))))..))).).)))))))))))))))..).-)))))))..).)).))..)))...... ( -51.00) >DroPse_CAF1 124734 99 - 1 CUGCGGCAAUGUGUGUGUC--UGUCCGGCUCUGGAGUGCGGA-----GAAUCGCCUACC----------CCCCGCCUCCAAGCUGCGAC-GGACACGUGUGAUAGUUGCAAUUUAUA .((((((.(((..((((((--(((((((((.(((((.((((.-----(...........----------).)))))))))))))).)))-)))))))..).)).))))))....... ( -46.70) >DroSim_CAF1 65642 114 - 1 UUGUGGCAAUGUGUGUGUC--UCUGCGGCUCCGGAGCCCGGACUCCAGAUCCGAUUACGGACGCGGAUGAUUCGACUCCGGGUCGCGGC-GGACACGUGUGAUAGUUGCAAUUUAUA (((..((.(((..((((((--((((((((.((((((..((((((((.(.((((....)))))..))).).)))).))))))))))))).-)))))))..).)).))..)))...... ( -52.50) >DroEre_CAF1 71474 114 - 1 UUGUGGCAAUGUGUGUGUC--UCUCCGGCUCCGGAGCCCGGACUCCAGAUCCGGCUACGGACGCGGAUGAUUCGACUCCGGGCCGCGGC-GGACACGUGUGAUAGUUGCAAUUUAUA (((..((.(((..((((((--(.((((((.((((((..((((((((.(.((((....)))))..))).).)))).)))))))))).)).-)))))))..).)).))..)))...... ( -51.20) >DroYak_CAF1 70275 114 - 1 UUGUGGCAAUGUGUGUGUC--UCUCCGGCUCCGGAGGCCGGACUCCAGAUCCGGAUAUGGACGCGGAUGAUUCGACUCCGGGUCGCGGC-GGACACGUGUGAUAGUUGCAAUUUAUA (((..((.(((..((((((--(.((((((.((((((..((((((((.(.((((....)))))..))).).)))).)))))))))).)).-)))))))..).)).))..)))...... ( -47.10) >DroAna_CAF1 73580 111 - 1 AUGUGGCAAUGAGUGUGUCUCUCUCCGGCUCCGGG--UCGGUCUACGGAUAA----UUGGAGUCUACGGAUUCGACUUAAAGUCGGGAUGGGACACGUGUGAUAGUUGCAAUUUAUA .((..((.(((..((((((((..(((((((..(((--((.(((....)))..----...(((((....))))))))))..)))))))..))))))))..).)).))..))....... ( -44.20) >consensus UUGUGGCAAUGUGUGUGUC__UCUCCGGCUCCGGAGCCCGGACUCCAGAUCCGGAUACGGACGCGGAUGAUUCGACUCCGGGUCGCGGC_GGACACGUGUGAUAGUUGCAAUUUAUA .((((((.(((..((((((...((.((((.((((((..((((.(((...((((....))))...)))...)))).)))))))))).))...))))))..).)).))))))....... (-29.45 = -31.33 + 1.88)

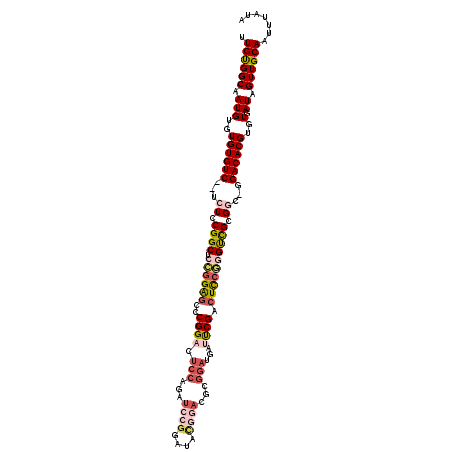
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:30 2006