| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,338,714 – 2,338,816 |
| Length | 102 |
| Max. P | 0.641694 |

| Location | 2,338,714 – 2,338,816 |
|---|---|
| Length | 102 |
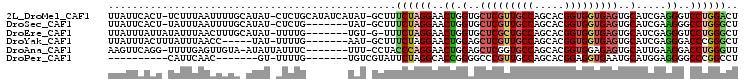
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 72.42 |
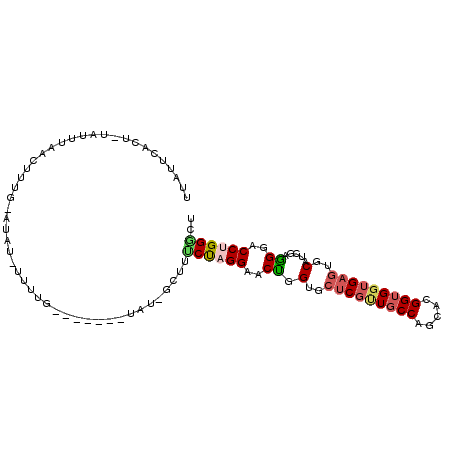
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -17.18 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2338714 102 + 22407834 UUAUUCACU-UCUUUAAUUUUGCAUAU-CUCUGCAUAUCAUAU-GCUUUCUAGGAACUGGUGCUCGUUGCCAGCACGGUGGUGAGUGCAUCGAGGGUCCUGGACU .........-...........((((((-...........))))-))..(((((((.((.(..((((..(((.....)))..))))..).....)).))))))).. ( -29.40) >DroSec_CAF1 45258 95 + 1 UUAUUCACU-UAUUUAAUUUUGCAUAU-CUCUG-------UAU-GCUUUCUAGGAACUGGUGCUCGUUGCCAGCACGGUGGUGAGUGCAUCGAAGGGCCUGGGCU .........-...........((((((-....)-------)))-))..((((((..((.(..((((..(((.....)))..))))..).....))..)))))).. ( -30.30) >DroEre_CAF1 50360 95 + 1 UUAUUUAUUAUAUUUAACUUUGCAUAU-UUUUG-------UGU-G-UUUCUAGGAACUGGUGCUCGCUGCCAGCACGGUGGUGAGUGCAUCGAGGGUCCUGGGCU .....................((((((-....)-------)))-)-).(((((((.((.(..(((((..((.....))..)))))..).....)).))))))).. ( -31.40) >DroYak_CAF1 47203 91 + 1 UUAUUUACUUUAUUUAACC-----UAU-UUUUG-------AAU-GCUUUCUAGGAACUGGAGCUCGUUGCCAGCACGGUGGUGAGUGCAUCGAGGGACCCGGGCU .................((-----(.(-(((((-------(.(-((..(((((...)))))(((((..(((.....)))..)))))))))))))))....))).. ( -21.90) >DroAna_CAF1 49179 95 + 1 AAGUUCAGG-UUUUGAGUUGUA-AUAUUAUUUC-------UUU-CCUACCCAGGAACUGGAGCUCGGUGCCAGCACGGUGGAGAGUGCAUUGAAGGACCUGGGUU .((..((((-((((....((((-.........(-------(((-((.(((..((.((((.....)))).)).....)))))))))))))....))))))))..)) ( -27.30) >DroPer_CAF1 104600 80 + 1 ----------CAUUCAAC-------GU-UUUUG-------UGUCGUAUUCUAGGCACCGGGGCCCGUUGCCAGCACGGAGGUGAAUGCAUGGAGGGGCCCGGCCU ----------........-------..-....(-------((((........)))))((((.(((..(((...(((....)))...)))....))).)))).... ( -28.00) >consensus UUAUUCACU_UAUUUAACUUUG_AUAU_UUUUG_______UAU_GCUUUCUAGGAACUGGUGCUCGUUGCCAGCACGGUGGUGAGUGCAUCGAGGGACCUGGGCU ................................................((((((..((.(..(((((((((.....)))))))))..).....))..)))))).. (-17.18 = -17.68 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:18 2006