
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,283,507 – 2,283,602 |
| Length | 95 |
| Max. P | 0.893665 |

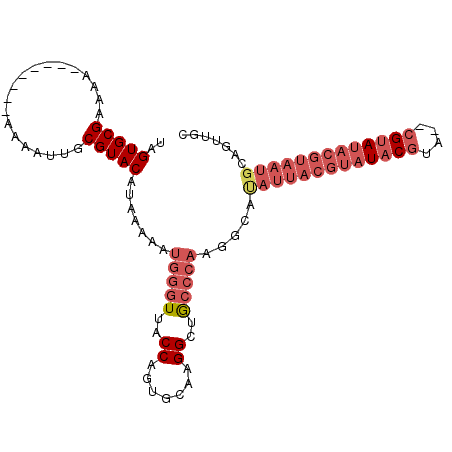
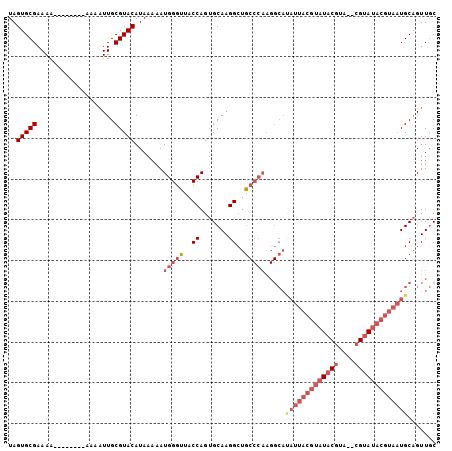
| Location | 2,283,507 – 2,283,602 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -21.43 |
| Energy contribution | -23.63 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2283507 95 + 22407834 UAGUGCGAAAA--------AAAAUUGCGUACAUAAAAAUGGGUUACCAGUGCAAGGCUGCCCAAGGCAUAUUACGUAUACGUA--CGUAUACGUAAUGCAGUUGC ..(((((....--------.......))))).......(((((..((.......))..)))))..((((..(((((((((...--.)))))))))))))...... ( -29.30) >DroSec_CAF1 19725 94 + 1 UAGUGCGAAAA---------AAAUUGCGUACAUAAAAAUGGGUUACCAGUGCAAGGCUGCCCAAGGCACAUUACGUAUACGUA--CGUAUACGUAAUGCAGUUGC ..(((((....---------......))))).......(((((..((.......))..))))).(((.((((((((((((...--.))))))))))))..))).. ( -30.40) >DroSim_CAF1 19813 95 + 1 UAGUGCGAAAA--------AAAAUUGCGUACAUAAAAAUGGGUUACCAGUGCAAGGCUGCCCAAGGCACAUUACGUAUACGUA--CGUAUACGUAAUGCAGUUGC ..(((((....--------.......))))).......(((((..((.......))..))))).(((.((((((((((((...--.))))))))))))..))).. ( -30.30) >DroEre_CAF1 19777 95 + 1 UAGUGCGAAAA--------AACGUUGCGUACAUAAAAAUGGGUUACCAGUGCAAGGCUGCCCAAGGCAUAUUACGUAUACGUA--CGUAUACGUAAUGCAGUUGC ..(((((....--------.......))))).......(((((..((.......))..)))))..((((..(((((((((...--.)))))))))))))...... ( -29.30) >DroYak_CAF1 20074 103 + 1 UAGUGCGAAAAAAACGUAAAACGUUGCGUACAUAAAAAUGGGUUACCAGUGCAAGGCUGCCCAAGGCAUAUUACGUAUACGUA--CGUAUACGUAAUGCAGUUGC ..(((((.....((((.....)))).))))).......(((((..((.......))..)))))..((((..(((((((((...--.)))))))))))))...... ( -33.90) >DroAna_CAF1 19414 79 + 1 UAGUGCGAAAAA-------AAAGUUGCGUACAUAAAAAUGGGUUACCAGAGCAAGGGUA------------------UACAUUUUCG-AUUUAAAUUGCGGUUUC ..(((((.((..-------....)).)))))...((((((...((((........))))------------------..))))))..-................. ( -11.90) >consensus UAGUGCGAAAA________AAAAUUGCGUACAUAAAAAUGGGUUACCAGUGCAAGGCUGCCCAAGGCAUAUUACGUAUACGUA__CGUAUACGUAAUGCAGUUGC ..(((((...................))))).......(((((..((.......))..))))).....(((((((((((((....)))))))))))))....... (-21.43 = -23.63 + 2.19)

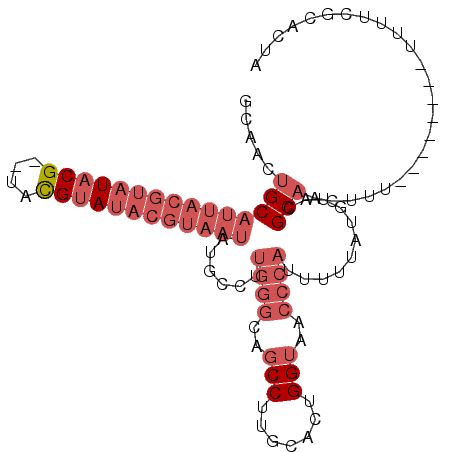
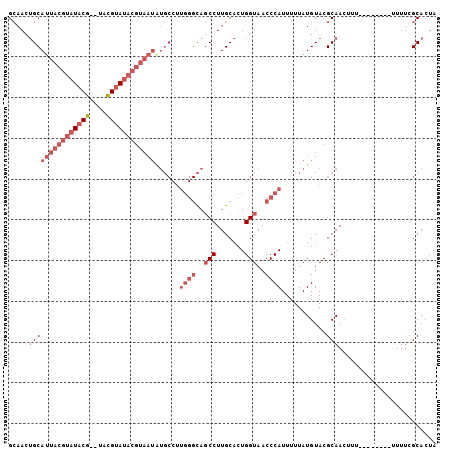
| Location | 2,283,507 – 2,283,602 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -17.45 |
| Energy contribution | -19.82 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2283507 95 - 22407834 GCAACUGCAUUACGUAUACG--UACGUAUACGUAAUAUGCCUUGGGCAGCCUUGCACUGGUAACCCAUUUUUAUGUACGCAAUUUU--------UUUUCGCACUA ((....(((((((((((((.--...)))))))))..))))..((((..(((.......)))..))))...........))......--------........... ( -25.50) >DroSec_CAF1 19725 94 - 1 GCAACUGCAUUACGUAUACG--UACGUAUACGUAAUGUGCCUUGGGCAGCCUUGCACUGGUAACCCAUUUUUAUGUACGCAAUUU---------UUUUCGCACUA ((....(((((((((((((.--...)))))))))))))....((((..(((.......)))..))))...........)).....---------........... ( -28.90) >DroSim_CAF1 19813 95 - 1 GCAACUGCAUUACGUAUACG--UACGUAUACGUAAUGUGCCUUGGGCAGCCUUGCACUGGUAACCCAUUUUUAUGUACGCAAUUUU--------UUUUCGCACUA ((....(((((((((((((.--...)))))))))))))....((((..(((.......)))..))))...........))......--------........... ( -28.90) >DroEre_CAF1 19777 95 - 1 GCAACUGCAUUACGUAUACG--UACGUAUACGUAAUAUGCCUUGGGCAGCCUUGCACUGGUAACCCAUUUUUAUGUACGCAACGUU--------UUUUCGCACUA ((...((((((((((((((.--...)))))))))))......((((..(((.......)))..))))...........)))..(..--------....))).... ( -25.80) >DroYak_CAF1 20074 103 - 1 GCAACUGCAUUACGUAUACG--UACGUAUACGUAAUAUGCCUUGGGCAGCCUUGCACUGGUAACCCAUUUUUAUGUACGCAACGUUUUACGUUUUUUUCGCACUA ((....(((((((((((((.--...)))))))))))......((((..(((.......)))..))))...........))((((.....))))......)).... ( -28.60) >DroAna_CAF1 19414 79 - 1 GAAACCGCAAUUUAAAU-CGAAAAUGUA------------------UACCCUUGCUCUGGUAACCCAUUUUUAUGUACGCAACUUU-------UUUUUCGCACUA ......((.........-.(((((((..------------------((((........))))...))))))).((....)).....-------......)).... ( -8.80) >consensus GCAACUGCAUUACGUAUACG__UACGUAUACGUAAUAUGCCUUGGGCAGCCUUGCACUGGUAACCCAUUUUUAUGUACGCAACUUU________UUUUCGCACUA .....(((((((((((((((....))))))))))))......((((..(((.......)))..))))...........)))........................ (-17.45 = -19.82 + 2.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:02 2006