| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 30,129 – 30,231 |
| Length | 102 |
| Max. P | 0.751936 |

| Location | 30,129 – 30,231 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.44 |
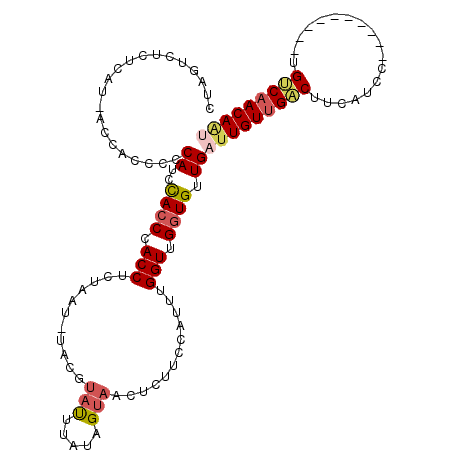
| Mean single sequence MFE | -16.75 |
| Consensus MFE | -12.97 |
| Energy contribution | -13.55 |
| Covariance contribution | 0.59 |
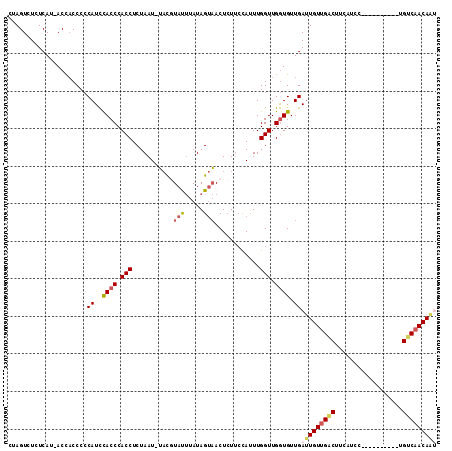
| Combinations/Pair | 1.19 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
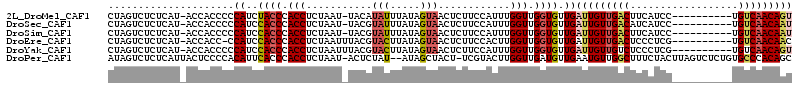
>2L_DroMel_CAF1 30129 102 - 22407834 CUAGUCUCUCAU-ACCACCCCCAUCUACCCACCUCUAAU-UACAUAUUUAUAGUAACUCUUCCAUUUGGUUGGUGUUGAUUGUUGACUUCAUCC----------UGUCAACAGU ........((((-((((.((..............(((..-..........)))..............)).))))).)))((((((((.......----------.)))))))). ( -16.49) >DroSec_CAF1 8039 102 - 1 CUAGUCUCUCAU-ACCACCCCCAUCCACCCACCUCUAAU-UACGUAUUUAUAGUAACUCUUCCAUUUGGUUGGUGUUGAUUGUUGACAUCAUCC----------UGUCAACAAU ............-........((..((((.(((......-....(((.....)))............))).)))).))((((((((((......----------)))))))))) ( -18.25) >DroSim_CAF1 9336 102 - 1 CUAGUCUCUCAU-ACCACCCCCAUCCACCCACCUCUAAU-UACGUAUUUAUAGUAACUCUUCCAUUUGGUUGGUGUUGAUUGUUGACUUCAUCC----------UGUCAACAAU ............-........((..((((.(((......-....(((.....)))............))).)))).))(((((((((.......----------.))))))))) ( -17.65) >DroEre_CAF1 8123 102 - 1 CUAGUCUCUCAU-ACCACC-CCAUCCACCCACCUCUAAUUUACGUACUUAUAGUAACUCUUCCACUUGGUUGGUGUUGAUUGUUGACUCCCUCG----------UGUCAACAAC ............-......-.((..((((.(((...........(((.....)))............))).)))).)).((((((((.......----------.)))))))). ( -19.30) >DroYak_CAF1 7985 103 - 1 CUAGUCUCUCAU-ACCACCCCCAUCCACCCACCUCUAAUUUACGUACUUAUAGUAACUCUUCCAUUUGGUUGGUGUUGAUUGUUGUCUCCCUCG----------UGUCAACAGU ............-........((..((((.(((...........(((.....)))............))).)))).))(((((((.(.......----------.).))))))) ( -16.10) >DroPer_CAF1 8035 110 - 1 AUAGUCUCUCAUUACUCCCCACAUUCACCCACCUCUAAU-ACUCUAU--AUAGCUACU-UCGUACUUGGUUGAUGUUGAAUGUUGGCUUUCUACUUAGUCUCUGUGCCCACAGC ..((.((..........((.(((((((..(((..(((((-((.....--.........-..))).))))..).)).))))))).))..........)).))((((....)))). ( -12.74) >consensus CUAGUCUCUCAU_ACCACCCCCAUCCACCCACCUCUAAU_UACGUAUUUAUAGUAACUCUUCCAUUUGGUUGGUGUUGAUUGUUGACUUCAUCC__________UGUCAACAAU .....................((..((((.(((...........(((.....)))............))).)))).))(((((((((..................))))))))) (-12.97 = -13.55 + 0.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:07 2006