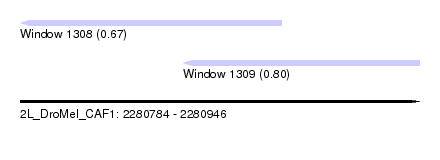
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,280,784 – 2,280,946 |
| Length | 162 |
| Max. P | 0.804859 |

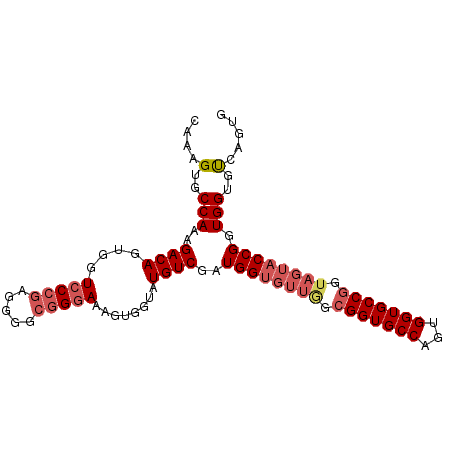
| Location | 2,280,784 – 2,280,890 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.73 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -30.91 |
| Energy contribution | -31.35 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2280784 106 - 22407834 AGCGGUGCCAGUGGUGCCAGUAGUACCGUUGGUGUCAGUGGGUCAGGCCC-UGGG---UGAGUGGGUGUUGCUAUAAUGGCAAAUCACUUUGCACCGCAAACAGAGUCCA .(((((((.((((((((((((((((((((((....)))))).....((((-....---.....))))..))))))..)))))..)))))..)))))))............ ( -39.60) >DroSec_CAF1 16974 106 - 1 GGCGGUGCCAGUGGUGCCGGUAGUACCGGUGGUGUCAGUGGGUCAGGCCC-UGGG---UGAGUGGGUGUUGCUAUAAUGGCAAAUCACUUUGCACCGCAAACAGAGUCCA .(((((((.(((((((((((.....)))))..(((((...(((.(.((((-....---.....)))).).)))....))))).))))))..)))))))............ ( -43.70) >DroSim_CAF1 17080 106 - 1 GGCGGUGCCAGUGGUGCCGGUAGUACCGGUGGUGUCAGUGGGUCAGGCCC-UGGG---UGAGUGGGUGUUGCUAUAAUGGCAAAUCACUUUGCACCGCAAACAGAGUCCA .(((((((.(((((((((((.....)))))..(((((...(((.(.((((-....---.....)))).).)))....))))).))))))..)))))))............ ( -43.70) >DroYak_CAF1 17383 95 - 1 GGCGGUGUUG---------------GCGGUGGUGUCAGUGGGUCAGGCCCCUGGGUACUGGAUGGGUGGUGCUAUAAUGGCAAAUCACUUUGCACCGCAAACAGAGUCCA (((..((((.---------------(((((((..(((.(.(..((((((....))).)))..).).)))..))......(((((....)))))))))).))))..))).. ( -35.00) >consensus GGCGGUGCCAGUGGUGCCGGUAGUACCGGUGGUGUCAGUGGGUCAGGCCC_UGGG___UGAGUGGGUGUUGCUAUAAUGGCAAAUCACUUUGCACCGCAAACAGAGUCCA .(((((((((.((((((.....)))))).)).(((((...(((.(.((((.(.........).)))).).)))....))))).........)))))))............ (-30.91 = -31.35 + 0.44)

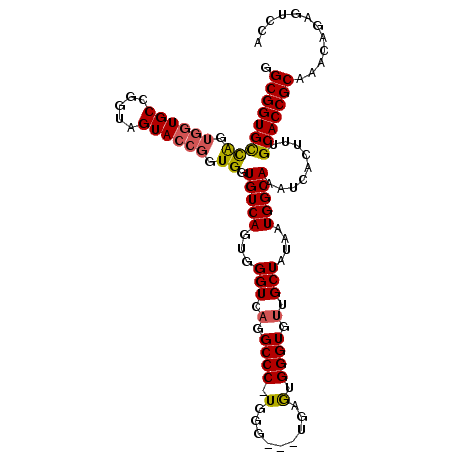
| Location | 2,280,850 – 2,280,946 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -23.02 |
| Energy contribution | -25.40 |
| Covariance contribution | 2.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2280850 96 - 22407834 CAAAGUGCCAAAGACAGUGGUCCCGAGGGGCGGGAAAGUGGUAUGUCGAUGGUGUUAGCGGUGCCAGUGGUGCCAGUAGUACCGUUGGUGUCAGUG ...((..(((..((((....(((((.....)))))........))))..)))..))...((..((((((((((.....))))))))))..)).... ( -36.40) >DroSec_CAF1 17040 96 - 1 CAAAGUGCCAAAGACAGUGGUCCCGAGGGGCGGGAAAGUGGUAUGUCGAUGGUGUUGGCGGUGCCAGUGGUGCCGGUAGUACCGGUGGUGUCAGUG ...((..(((..((((....(((((.....)))))........))))..)))..))...((..(((.((((((.....)))))).)))..)).... ( -37.50) >DroSim_CAF1 17146 96 - 1 CAAAGUGCCAAAGACAGUGGUCCCGAGGGGCGGGAAAGUGGUAUGUCGAUGGUGUUGGCGGUGCCAGUGGUGCCGGUAGUACCGGUGGUGUCAGUG ...((..(((..((((....(((((.....)))))........))))..)))..))...((..(((.((((((.....)))))).)))..)).... ( -37.50) >DroEre_CAF1 17169 72 - 1 CAAAGUGCCAAAGACAGUGGUCCCGGGGGGAAGUAAAGUGGUAUGUCGAUGGUGGUU------------------------CCGGUGGUGCCGGUG ..((.(((((..((((....((((...))))............))))..))))).))------------------------((((.....)))).. ( -18.89) >consensus CAAAGUGCCAAAGACAGUGGUCCCGAGGGGCGGGAAAGUGGUAUGUCGAUGGUGUUGGCGGUGCCAGUGGUGCCGGUAGUACCGGUGGUGUCAGUG ....(..(((..((((....(((((.....)))))........))))..((((((((.(((((((...))))))).)))))))).)))..)..... (-23.02 = -25.40 + 2.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:57 2006