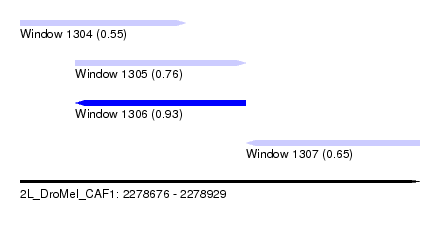
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,278,676 – 2,278,929 |
| Length | 253 |
| Max. P | 0.931677 |

| Location | 2,278,676 – 2,278,781 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.37 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -27.67 |
| Energy contribution | -30.12 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
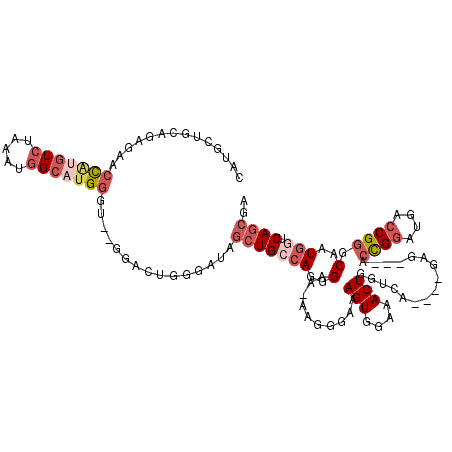
>2L_DroMel_CAF1 2278676 105 + 22407834 AGC--CGCAUUUGUUGGGGU-UUUGGUUCAAGUGGGUCUCAUUGAGCCACUUCUAUGGCUGCCGCACCACCAUCGCCCCAGCAUGUGUCCCACAUUCUGUGGCUCUUC (((--((((..(((((((((-..((((..((((((.((.....)).)))))).....((....))...))))..)))))))))((((...))))...))))))).... ( -43.50) >DroSec_CAF1 15044 105 + 1 AGC--CGCAUUUGUUGGGGU-UUUGGUUCAAGUGGGUCUCAUUGAGCCACUUCUAUGGCUGCCGCACCACCAUCGCCCCAGCAUGUGUCCCACAUUCUGUGGCUCUUC (((--((((..(((((((((-..((((..((((((.((.....)).)))))).....((....))...))))..)))))))))((((...))))...))))))).... ( -43.50) >DroSim_CAF1 14995 105 + 1 AGC--CGCAUUUGUUGGGGU-UUUGGUUCAAGUGGGUCUCAUUGAGCCACUUCUAUGGCUGCCGCACCACCGUCGCCCCAGCAUGUGUCCCACAUUCUGUGGCUCCUC (((--((((..(((((((((-..((((..((((((.((.....)).)))))).....((....))...))))..)))))))))((((...))))...))))))).... ( -42.80) >DroEre_CAF1 15024 105 + 1 AGC--CGCAUUUGUUGGGGU-UUUGGUUCAAGUGGGUCUCAUUGAGCCACUUCUAUGGCUGCCGCGCCACCGCCUCCCCAGCAUGUGUUCCACAUUCUGUGGCUCUAC (((--((((..((((((((.-..((((..((((((.((.....)).))))))....(((......)))))))...))))))))((((...))))...))))))).... ( -43.50) >DroYak_CAF1 15324 93 + 1 AGC--CGCAUUUGUUGGGGU-UUUGGUUCAAGUGGGUCUCAUUGAGCCACUUCUAUGGCUGCC-----ACCACC-------CAUGUGUCCCACAUUCUGUGGCUCUUC (((--((((.......((((-..((((((((((((.((.....)).)))))....)))..)))-----)..)))-------)(((((...)))))..))))))).... ( -32.30) >DroAna_CAF1 14821 98 + 1 AGCUCGGCAUUUGUUGGGGCUUUCGGUUCAAGUGGGUCUCAUUGAGCCACUUCUAUGGCUGCUC-------CUCGCC---GCAUUUGGCCCACAUUUUGUGGCCCUCC ....((((.......(((((..(((....((((((.((.....)).))))))...)))..))))-------)..)))---).....((.((((.....)))).))... ( -33.00) >consensus AGC__CGCAUUUGUUGGGGU_UUUGGUUCAAGUGGGUCUCAUUGAGCCACUUCUAUGGCUGCCGCACCACCAUCGCCCCAGCAUGUGUCCCACAUUCUGUGGCUCUUC ......((((.(((((((((...((((..((((((.((.....)).))))))....((...)).....))))..))))))))).)))).((((.....))))...... (-27.67 = -30.12 + 2.44)

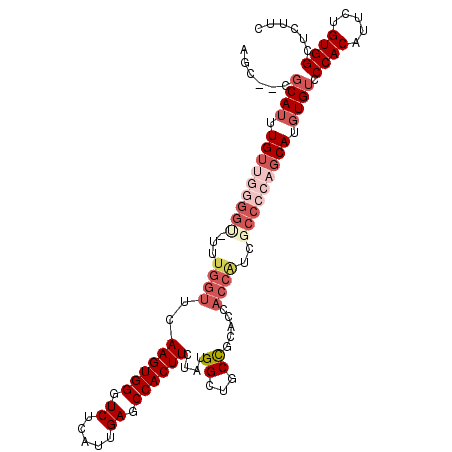
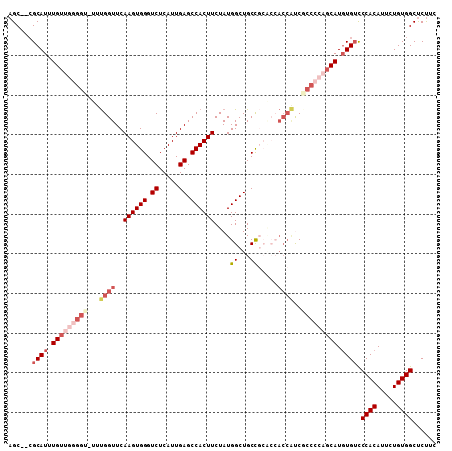
| Location | 2,278,711 – 2,278,819 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -13.40 |
| Energy contribution | -14.76 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
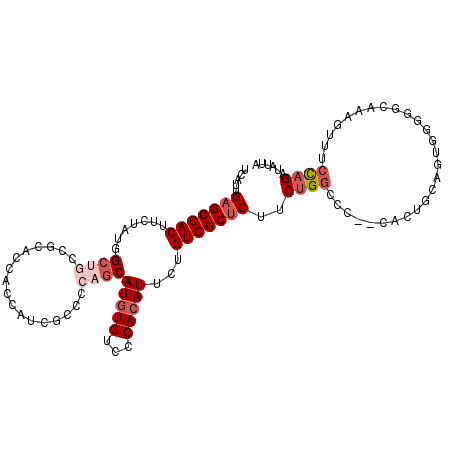
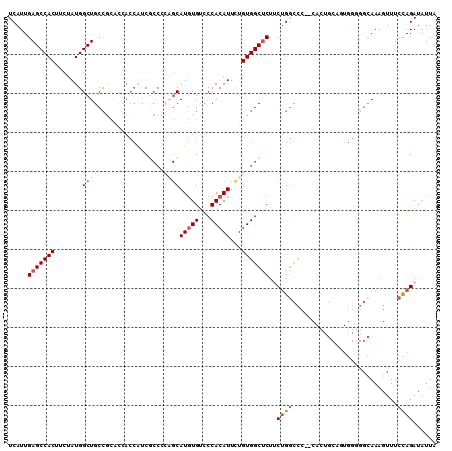
>2L_DroMel_CAF1 2278711 108 + 22407834 UCAUUGAGCCACUUCUAUGGCUGCCGCACCACCAUCGCCCCAGCAUGUGUCCCACAUUCUGUGGCUCUUCUGGCCC--CACUGCAGUGGGGGCAAAGUUUCCAGAUAUUA .....(((((((.......((((..((.........))..))))(((((...)))))...))))))).((((((((--(((....)))))).........)))))..... ( -37.20) >DroSec_CAF1 15079 94 + 1 UCAUUGAGCCACUUCUAUGGCUGCCGCACCACCAUCGCCCCAGCAUGUGUCCCACAUUCUGUGGCUCUUCUGGUCC--C--------------AAAGUUUCCAGAUAUUA .....(((((((.......((((..((.........))..))))(((((...)))))...))))))).(((((...--.--------------.......)))))..... ( -26.60) >DroSim_CAF1 15030 108 + 1 UCAUUGAGCCACUUCUAUGGCUGCCGCACCACCGUCGCCCCAGCAUGUGUCCCACAUUCUGUGGCUCCUCUGGCCC--CACAGCUAUGGGGGCAAAGUUUCCAGAUAUUA .....(((((((.......((((..((((....)).))..))))(((((...)))))...))))))).((((((((--(.((....)))))))........))))..... ( -34.30) >DroEre_CAF1 15059 108 + 1 UCAUUGAGCCACUUCUAUGGCUGCCGCGCCACCGCCUCCCCAGCAUGUGUUCCACAUUCUGUGGCUCUACUGGCCC--CACUGCAGUGGGGGCAAAGUUUCCAGAUAUUA .....(((((((.....((((......))))..........((.(((((...))))).)))))))))...((.(((--(((....)))))).))................ ( -36.60) >DroYak_CAF1 15359 98 + 1 UCAUUGAGCCACUUCUAUGGCUGCC-----ACCACC-------CAUGUGUCCCACAUUCUGUGGCUCUUCUGGCCCACCAAUGCAGUGGUGGCAAAGUUUCCAGAUAUUA ...((((((((......)))))(((-----(((((.-------.(((((...)))))...((((..(....)..)))).......))))))))........)))...... ( -30.00) >DroAna_CAF1 14859 83 + 1 UCAUUGAGCCACUUCUAUGGCUGCUC-------CUCGCC---GCAUUUGGCCCACAUUUUGUGGCCCUCCCGUC-----------------GCAAACUUUCUAGAUAUUA ......(((((......)))))....-------...((.---((....((.((((.....)))).))....)).-----------------))................. ( -18.10) >consensus UCAUUGAGCCACUUCUAUGGCUGCCGCACCACCAUCGCCCCAGCAUGUGUCCCACAUUCUGUGGCUCUUCUGGCCC__CACUGCAGUGGGGGCAAAGUUUCCAGAUAUUA .....(((((((.......((((.................))))(((((...)))))...)))))))..((((...........................))))...... (-13.40 = -14.76 + 1.36)
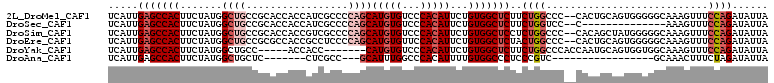
| Location | 2,278,711 – 2,278,819 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -21.47 |
| Energy contribution | -22.29 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
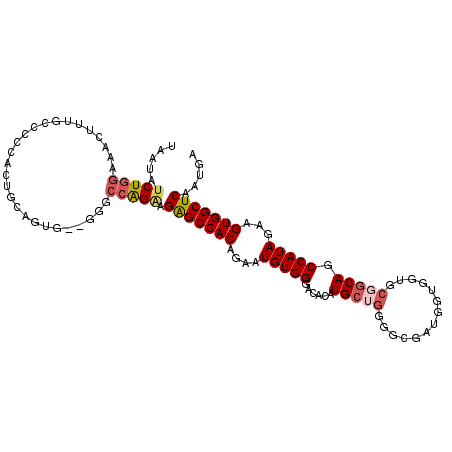
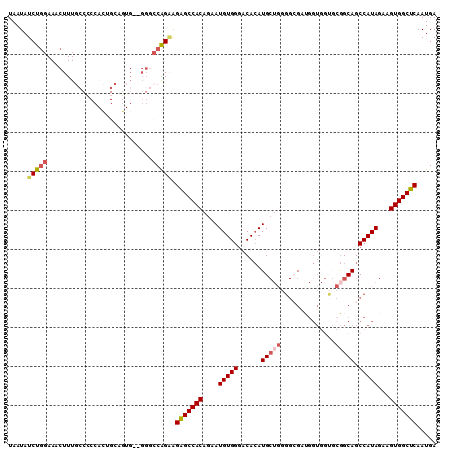
>2L_DroMel_CAF1 2278711 108 - 22407834 UAAUAUCUGGAAACUUUGCCCCCACUGCAGUG--GGGCCAGAAGAGCCACAGAAUGUGGGACACAUGCUGGGGCGAUGGUGGUGCGGCAGCCAUAGAAGUGGCUCAAUGA .....(((((.........((((((....)))--)))))))).(((((((....(((((......(((((.(.((....)).).))))).)))))...)))))))..... ( -41.70) >DroSec_CAF1 15079 94 - 1 UAAUAUCUGGAAACUUU--------------G--GGACCAGAAGAGCCACAGAAUGUGGGACACAUGCUGGGGCGAUGGUGGUGCGGCAGCCAUAGAAGUGGCUCAAUGA .....(((((...(...--------------.--)..))))).(((((((....(((((......(((((.(.((....)).).))))).)))))...)))))))..... ( -31.20) >DroSim_CAF1 15030 108 - 1 UAAUAUCUGGAAACUUUGCCCCCAUAGCUGUG--GGGCCAGAGGAGCCACAGAAUGUGGGACACAUGCUGGGGCGACGGUGGUGCGGCAGCCAUAGAAGUGGCUCAAUGA .............(((((.((((((....)))--))).)))))(((((((....(((((......(((((.(.((....)).).))))).)))))...)))))))..... ( -41.30) >DroEre_CAF1 15059 108 - 1 UAAUAUCUGGAAACUUUGCCCCCACUGCAGUG--GGGCCAGUAGAGCCACAGAAUGUGGAACACAUGCUGGGGAGGCGGUGGCGCGGCAGCCAUAGAAGUGGCUCAAUGA ........(....).(((.((((((....)))--))).)))..(((((((...(((((...)))))(((.....))).(((((......)))))....)))))))..... ( -40.50) >DroYak_CAF1 15359 98 - 1 UAAUAUCUGGAAACUUUGCCACCACUGCAUUGGUGGGCCAGAAGAGCCACAGAAUGUGGGACACAUG-------GGUGGU-----GGCAGCCAUAGAAGUGGCUCAAUGA .....(((((........((((((......)))))).))))).(((((((...(((((...))))).-------.(((((-----....)))))....)))))))..... ( -36.10) >DroAna_CAF1 14859 83 - 1 UAAUAUCUAGAAAGUUUGC-----------------GACGGGAGGGCCACAAAAUGUGGGCCAAAUGC---GGCGAG-------GAGCAGCCAUAGAAGUGGCUCAAUGA .............((((.(-----------------(.((.(.((.((((.....)))).))...).)---).))..-------))))((((((....))))))...... ( -23.00) >consensus UAAUAUCUGGAAACUUUGCCCCCACUGCAGUG__GGGCCAGAAGAGCCACAGAAUGUGGGACACAUGCUGGGGCGAUGGUGGUGCGGCAGCCAUAGAAGUGGCUCAAUGA .....(((((...........................))))).(((((((....(((((......(((((..............))))).)))))...)))))))..... (-21.47 = -22.29 + 0.82)

| Location | 2,278,819 – 2,278,929 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -18.16 |
| Energy contribution | -20.07 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2278819 110 - 22407834 CAUUCCGCAGAGAACCAUGUCUAGA-GGCACGGGU--GGACUGGGAUAGCUGCCAGGGAAAAAGGGAAAGUGAAAACUGGUCA----GAG---ACCGGAUGACCGGGCAAUGGUCAGCGA ...(((.(((...(((.((((....-))))..)))--...))))))..(((((((..(......((..(((....)))..)).----...---.((((....)))).)..))).)))).. ( -35.40) >DroSec_CAF1 15173 110 - 1 CAUGCUGCAGAGAACCAUGUGUAGAUGGCAUGGUU--GGACUGGGAUUGCUGGCAGGGAA-AAGGGAAAGUGGAAACUGGUCA----GAG---ACCGGAUGACCGGGCAAUGGUCAGCGA .......(((..((((((((.......))))))))--...)))...((((((((......-...((..(((....)))..)).----...---.((((....))))......)))))))) ( -33.10) >DroSim_CAF1 15138 110 - 1 CAUGCUGCAGAGAACCAUGUCUAGAUGGCAUGGUU--GGACUGGGAUAGCUGCCAGGGAA-AAGGGAAAGUGGAAACUGGUCA----GAG---ACCGGAUGACCGGGCAAUGGUCAGCGA .......(((..(((((((((.....)))))))))--...))).....(((((((..(..-...((..(((....)))..)).----...---.((((....)))).)..))).)))).. ( -36.80) >DroEre_CAF1 15167 110 - 1 CAUGCUGCAGAGAACCAUGUCUAAAUGGCAUGGGU--GGACUGGGAUAGCUGCCAGGGAA-AAGGGAAAGUGGAAACUGGUCA----GAG---ACCGGGUGACCGGGCAAUGGUCAGCGA .......(((....(((((((.....)))))))..--...))).....(((((((..(..-...((..(((....)))..)).----...---.((((....)))).)..))).)))).. ( -39.20) >DroYak_CAF1 15457 114 - 1 CAUGCUGCAGACAAGCAUGUCUAAAUGGCCUGGGU--GGACUGGGAUGGCUGCCAGGGAA-AAGCAAAAGUGGAAACUGGACACAGAGAG---CCCGGGUGACCGGGCAAUGGUCAGCGA ((((((.......)))))).........((..(..--...)..))...(((((((.....-...............(((....)))...(---(((((....))))))..))).)))).. ( -42.80) >DroAna_CAF1 14942 94 - 1 CAUGCUGCUGCU-GCUGCCUCCAAAUGGCAUGGGUGUGGACUG------AUGAGAGGGGU-GGG--------------GGUCA----GAAAAGAGUGACCGUCCGGGCAAUGGUCAGAGA ....((.(((((-..(((((((...(..(((.(((....))).------)))..).))..-(((--------------(((((----........))))).))))))))..)).))))). ( -28.80) >consensus CAUGCUGCAGAGAACCAUGUCUAAAUGGCAUGGGU__GGACUGGGAUAGCUGCCAGGGAA_AAGGGAAAGUGGAAACUGGUCA____GAG___ACCGGAUGACCGGGCAAUGGUCAGCGA ..............(((((((.....)))))))...............(((((((..(..........(((....)))................((((....)))).)..))).)))).. (-18.16 = -20.07 + 1.90)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:55 2006