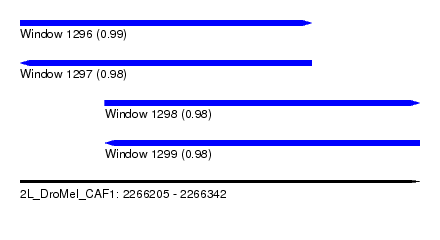
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,266,205 – 2,266,342 |
| Length | 137 |
| Max. P | 0.986930 |

| Location | 2,266,205 – 2,266,305 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.93 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.82 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
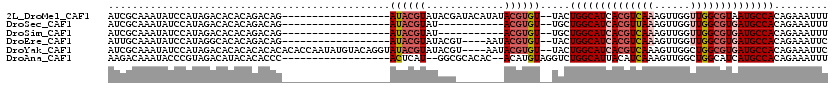
>2L_DroMel_CAF1 2266205 100 + 22407834 AUCGCAAAUAUCCAUAGACACACAGACAG------------------AUACGUAUACGAUACAUAUACGUGU--UACUGGCAUCACGUCAAAGUUGGUUGGCGUAAUGCCACAGAAAUUU .((.........................(------------------(((((((((.......)))))))))--)..((((((.(((((((......))))))).))))))..))..... ( -26.10) >DroSec_CAF1 2592 89 + 1 AUCGCAAAUAUCCAUAGACACACAGACAG------------------AUACGUAU-----------ACGUGU--UGCUGGCAUCACGUUAAAGUUGGUUGGCGUGAUGCCACAGAAAUUU .((....................((...(------------------(((((...-----------.)))))--).))(((((((((((((......)))))))))))))...))..... ( -26.80) >DroSim_CAF1 2613 89 + 1 AUCGCAAAUAUCCAUAGACACACAGACAG------------------AUACGUAU-----------ACGUGU--UGCUGGCAUCACGUCAAAGUUGGUUGGCGUGAUGCCACAGAAAUUU .((....................((...(------------------(((((...-----------.)))))--).))(((((((((((((......)))))))))))))...))..... ( -28.90) >DroEre_CAF1 2624 96 + 1 AUUGCAAAUAUCCAUAGGCACACAGACAG------------------AUACGUAUACGU----AAUACGUGU--UACUGGCAUCACGUCAAAGUUGGUUGGCGUGAUGCCACAGAAAUUC ..(((..((....))..)))........(------------------((((((((....----.))))))))--)..((((((((((((((......))))))))))))))......... ( -31.90) >DroYak_CAF1 2517 114 + 1 AUCGCAAAUAUCCAUAGACACACACACACACACCAAUAUGUACAGGUAUACGUAUACGU----AAUACGUGU--UACUGGCAUCACGUCAAAGUUGGCUGGCGUGAUGCCACAGAAAUUC .((.............(((((........(((......)))....(((((((....)))----.))))))))--)..(((((((((((((........)))))))))))))..))..... ( -32.40) >DroAna_CAF1 2871 98 + 1 AAGACAAAUACCCGUAGACAUACACACCC------------------ACUCAU--GGCGCACAC--ACAUGUAGGUCUGGCAUUACAUCAAAGUUGGCUGGCAUCAUGCCACAGAAAUUU .((((........(((....)))......------------------.(((((--(........--.)))).))))))(((...((......))..)))(((.....))).......... ( -16.70) >consensus AUCGCAAAUAUCCAUAGACACACAGACAG__________________AUACGUAUACG________ACGUGU__UACUGGCAUCACGUCAAAGUUGGUUGGCGUGAUGCCACAGAAAUUU ...............................................((((((.............)))))).....((((((((((((((......))))))))))))))......... (-19.60 = -20.82 + 1.22)
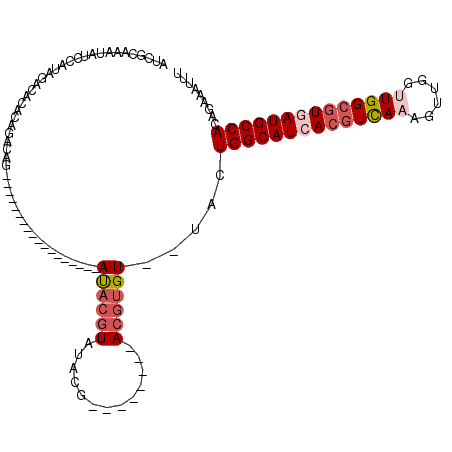
| Location | 2,266,205 – 2,266,305 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.93 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -19.62 |
| Energy contribution | -18.74 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2266205 100 - 22407834 AAAUUUCUGUGGCAUUACGCCAACCAACUUUGACGUGAUGCCAGUA--ACACGUAUAUGUAUCGUAUACGUAU------------------CUGUCUGUGUGUCUAUGGAUAUUUGCGAU .........((((((((((.(((......))).))))))))))(((--(.(((((((((...)))))))))..------------------.((((((((....)))))))).))))... ( -30.70) >DroSec_CAF1 2592 89 - 1 AAAUUUCUGUGGCAUCACGCCAACCAACUUUAACGUGAUGCCAGCA--ACACGU-----------AUACGUAU------------------CUGUCUGUGUGUCUAUGGAUAUUUGCGAU .......((((((((((((..............)))))))))).))--......-----------...((((.------------------.((((((((....))))))))..)))).. ( -26.04) >DroSim_CAF1 2613 89 - 1 AAAUUUCUGUGGCAUCACGCCAACCAACUUUGACGUGAUGCCAGCA--ACACGU-----------AUACGUAU------------------CUGUCUGUGUGUCUAUGGAUAUUUGCGAU .......((((((((((((.(((......))).)))))))))).))--......-----------...((((.------------------.((((((((....))))))))..)))).. ( -29.20) >DroEre_CAF1 2624 96 - 1 GAAUUUCUGUGGCAUCACGCCAACCAACUUUGACGUGAUGCCAGUA--ACACGUAUU----ACGUAUACGUAU------------------CUGUCUGUGUGCCUAUGGAUAUUUGCAAU .........((((((((((.(((......))).))))))))))(((--(.((((((.----....))))))..------------------.((((((((....)))))))).))))... ( -29.70) >DroYak_CAF1 2517 114 - 1 GAAUUUCUGUGGCAUCACGCCAGCCAACUUUGACGUGAUGCCAGUA--ACACGUAUU----ACGUAUACGUAUACCUGUACAUAUUGGUGUGUGUGUGUGUGUCUAUGGAUAUUUGCGAU .........((((((((((.(((......))).))))))))))(((--(..((((.(----(((((((((((((((..........))))))))))))))))..)))).....))))... ( -36.00) >DroAna_CAF1 2871 98 - 1 AAAUUUCUGUGGCAUGAUGCCAGCCAACUUUGAUGUAAUGCCAGACCUACAUGU--GUGUGCGCC--AUGAGU------------------GGGUGUGUAUGUCUACGGGUAUUUGUCUU ((((.(((((((((((......((..((......))...))((.((((((.(((--(.(....))--))).))------------------)))).)))))).))))))).))))..... ( -24.00) >consensus AAAUUUCUGUGGCAUCACGCCAACCAACUUUGACGUGAUGCCAGUA__ACACGU________CGUAUACGUAU__________________CUGUCUGUGUGUCUAUGGAUAUUUGCGAU ((((.((((((((((((((.(((......))).)))))))))........((((.............))))..................................))))).))))..... (-19.62 = -18.74 + -0.88)
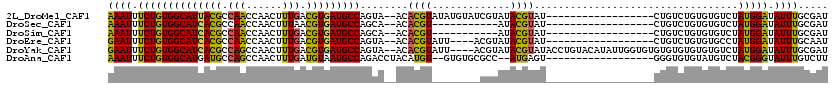
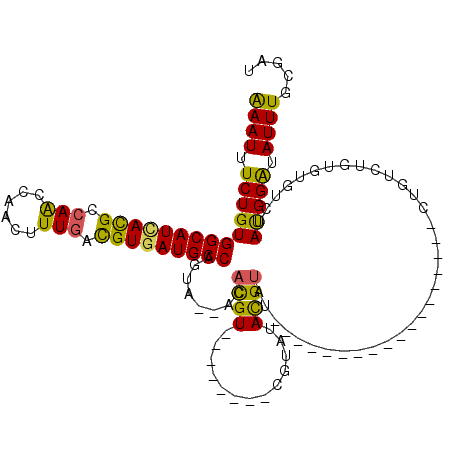
| Location | 2,266,234 – 2,266,342 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -19.87 |
| Energy contribution | -21.04 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
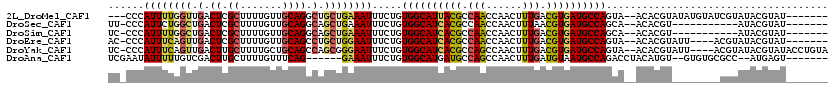
>2L_DroMel_CAF1 2266234 108 + 22407834 -------AUACGUAUACGAUACAUAUACGUGU--UACUGGCAUCACGUCAAAGUUGGUUGGCGUAAUGCCACAGAAAUUUCAGCAGCCUGCAACAAAAGCGAGUCAACCAAAAUGGG--- -------(((((((((.......)))))))))--...((((((.(((((((......))))))).))))))..............((.(((.......))).))...((.....)).--- ( -26.60) >DroSec_CAF1 2621 99 + 1 -------AUACGUAU-----------ACGUGU--UGCUGGCAUCACGUUAAAGUUGGUUGGCGUGAUGCCACAGAAAUUUCAGCUGCCUGCAACAAAAGCGAGUCAGCCAGAAUGGG-AA -------(((((...-----------.)))))--((.((((((((((((((......))))))))))))))))...((((..((((.((((.......)).)).))))..))))...-.. ( -33.70) >DroSim_CAF1 2642 99 + 1 -------AUACGUAU-----------ACGUGU--UGCUGGCAUCACGUCAAAGUUGGUUGGCGUGAUGCCACAGAAAUUUCAGCUGCCUGCAACAAAAGCGAGUCAGCCAAAAUGGG-GA -------(((((...-----------.)))))--((.((((((((((((((......))))))))))))))))...((((..((((.((((.......)).)).))))..))))...-.. ( -35.40) >DroEre_CAF1 2653 106 + 1 -------AUACGUAUACGU----AAUACGUGU--UACUGGCAUCACGUCAAAGUUGGUUGGCGUGAUGCCACAGAAAUUCCAGCAGGCUGCAACAAAAGCGAGUCAACUGAAAUGGG-GU -------((((((((....----.))))))))--...((((((((((((((......)))))))))))))).....((((((.((((((((.......)).)))...)))...))))-)) ( -39.00) >DroYak_CAF1 2557 113 + 1 UACAGGUAUACGUAUACGU----AAUACGUGU--UACUGGCAUCACGUCAAAGUUGGCUGGCGUGAUGCCACAGAAAUUCCCGCUGGCUGCAGCAAAAGCAAGUCAACUGAAAUGGG-GA ....(((((((((((....----.))))))).--))))((((((((((((........))))))))))))........((((((((....)))).....((..((....))..))))-)) ( -42.40) >DroAna_CAF1 2900 103 + 1 -------ACUCAU--GGCGCACAC--ACAUGUAGGUCUGGCAUUACAUCAAAGUUGGCUGGCAUCAUGCCACAGAAAUUUC------CUGAAACAAAAGCAAGUCGACAAAAAUAUUCGA -------.(((((--(........--.)))).))..................((((((((((.....))).(((.......------)))...........)))))))............ ( -18.60) >consensus _______AUACGUAUACG________ACGUGU__UACUGGCAUCACGUCAAAGUUGGUUGGCGUGAUGCCACAGAAAUUUCAGCUGCCUGCAACAAAAGCGAGUCAACCAAAAUGGG_GA .......((((((.............)))))).....((((((((((((((......)))))))))))))).................(((.......)))................... (-19.87 = -21.04 + 1.17)

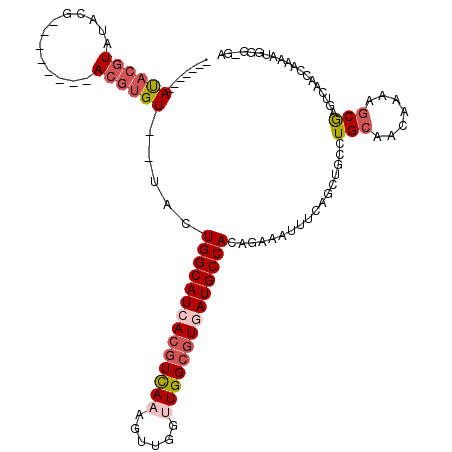
| Location | 2,266,234 – 2,266,342 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -30.81 |
| Consensus MFE | -21.78 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
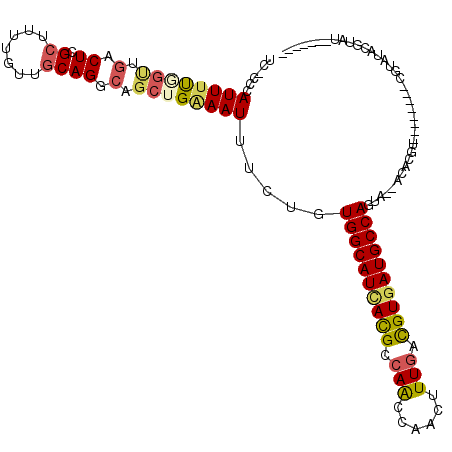
>2L_DroMel_CAF1 2266234 108 - 22407834 ---CCCAUUUUGGUUGACUCGCUUUUGUUGCAGGCUGCUGAAAUUUCUGUGGCAUUACGCCAACCAACUUUGACGUGAUGCCAGUA--ACACGUAUAUGUAUCGUAUACGUAU------- ---...((((..((.(.(..((.......))..)).))..)))).....((((((((((.(((......))).))))))))))...--..(((((((((...)))))))))..------- ( -32.80) >DroSec_CAF1 2621 99 - 1 UU-CCCAUUCUGGCUGACUCGCUUUUGUUGCAGGCAGCUGAAAUUUCUGUGGCAUCACGCCAACCAACUUUAACGUGAUGCCAGCA--ACACGU-----------AUACGUAU------- ..-....(((.(((((.(..((.......))..))))))))).....((((((((((((..............)))))))))).))--..(((.-----------...)))..------- ( -29.04) >DroSim_CAF1 2642 99 - 1 UC-CCCAUUUUGGCUGACUCGCUUUUGUUGCAGGCAGCUGAAAUUUCUGUGGCAUCACGCCAACCAACUUUGACGUGAUGCCAGCA--ACACGU-----------AUACGUAU------- ..-...((((..((((.(..((.......))..)))))..))))...((((((((((((.(((......))).)))))))))).))--..(((.-----------...)))..------- ( -34.90) >DroEre_CAF1 2653 106 - 1 AC-CCCAUUUCAGUUGACUCGCUUUUGUUGCAGCCUGCUGGAAUUUCUGUGGCAUCACGCCAACCAACUUUGACGUGAUGCCAGUA--ACACGUAUU----ACGUAUACGUAU------- ..-...((((((((.(.((.((.......)))).).)))))))).....((((((((((.(((......))).))))))))))...--..((((((.----....))))))..------- ( -33.70) >DroYak_CAF1 2557 113 - 1 UC-CCCAUUUCAGUUGACUUGCUUUUGCUGCAGCCAGCGGGAAUUUCUGUGGCAUCACGCCAGCCAACUUUGACGUGAUGCCAGUA--ACACGUAUU----ACGUAUACGUAUACCUGUA ((-((((..((....))..)).....((((....)))))))).......((((((((((.(((......))).))))))))))...--(((.((((.----(((....))))))).))). ( -35.60) >DroAna_CAF1 2900 103 - 1 UCGAAUAUUUUUGUCGACUUGCUUUUGUUUCAG------GAAAUUUCUGUGGCAUGAUGCCAGCCAACUUUGAUGUAAUGCCAGACCUACAUGU--GUGUGCGCC--AUGAGU------- ..(((..((((((..(((........))).)))------)))..)))..((((((.((..(((......)))..)).))))))......((((.--((....)))--)))...------- ( -18.80) >consensus UC_CCCAUUUUGGUUGACUCGCUUUUGUUGCAGGCAGCUGAAAUUUCUGUGGCAUCACGCCAACCAACUUUGACGUGAUGCCAGUA__ACACGU________CGUAUACGUAU_______ ......((((((((.(.((.((.......)))).).)))))))).....((((((((((.(((......))).))))))))))..................................... (-21.78 = -22.03 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:47 2006