| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,262,971 – 2,263,107 |
| Length | 136 |
| Max. P | 0.996848 |

| Location | 2,262,971 – 2,263,074 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.88 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -25.64 |
| Energy contribution | -25.32 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2262971 103 + 22407834 ----CGGGCGAUUGAUAAGGGGCCUCCAUGUCGGCCAA--CUCGCAUUAUCCAGCUCGAUUCAGUCGGCAAUUAAUGCCACUCCAAUUGCCCGAUUUGCCUGCACCUAC---------- ----((((((((((......((((........))))..--...((((((....((.((((...))))))...)))))).....))))))))))................---------- ( -34.70) >DroSec_CAF1 23518 113 + 1 ----CGGGCGUUUGAUAAGCGGCCUCCAUGUCGGCCAA--CUCGCAUUAUCCAGGCCGAUUCAGUCGGCAAUUAAUGCCACUCCAAUUGCCCGAUUUGCCUGCACCUCCGAAGGGGGUA ----((((((.(((......((((........))))..--...((((((.....((((((...))))))...)))))).....))).))))))..........(((.((....))))). ( -42.50) >DroSim_CAF1 24658 113 + 1 ----CGGGCGUUUGAUAAGCGGCCUCCAUGUCGGCCAA--CUCGCAUUAUCCAGCCCGAUUCAGUCGGCAAUUAAUGCCACUCCAAUUGCCCGAUUUGCCUGCACCUCCGAAUGGGGUA ----(((((....(((((((((((........))))..--...)).)))))..)))))....(((.((((.....))))))).....(((((.(((((..........))))).))))) ( -36.50) >DroEre_CAF1 25228 99 + 1 ----CGGGCGUUUGAUAAGCGGCCUCCAUGUUGGCCAA--CUCCCAUUAUCCAGCCCGUUUCAGCCGGCAAUUAAUGGCACUCCAAUUGCCCGAUUUGCCUGUAC-------------- ----((((((.(((......((((........))))..--...((((((....(((.((....)).)))...)))))).....))).))))))............-------------- ( -31.90) >DroYak_CAF1 31542 119 + 1 GUUGCGGGCGUUUGAUAAGCGCCCUCCAUGUGGGCCAACACUCACAUUAUCCAGCCCGAUUCAGGCGGCAAUUAAUGCCACUCCAAUUGCUUGAUUUGCCUGUACCUCGGAAUGAGGUA .....(((((((.....)))))))...(((((((......)))))))((((((..((((..(((((((((.....))))....(((....)))....)))))....))))..)).)))) ( -41.00) >consensus ____CGGGCGUUUGAUAAGCGGCCUCCAUGUCGGCCAA__CUCGCAUUAUCCAGCCCGAUUCAGUCGGCAAUUAAUGCCACUCCAAUUGCCCGAUUUGCCUGCACCUCCGAA_G_GGUA ....((((((.(((......((((........)))).......((((((....(.(((.......))))...)))))).....))).)))))).......................... (-25.64 = -25.32 + -0.32)

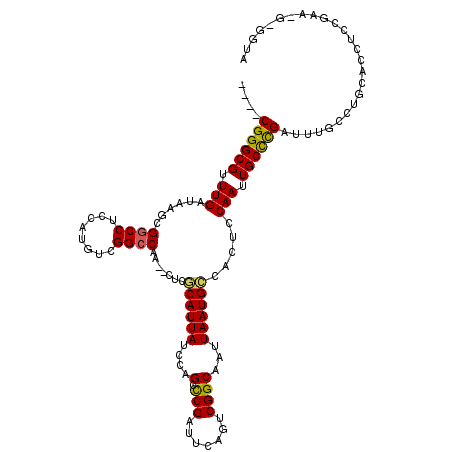
| Location | 2,263,005 – 2,263,107 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -21.37 |
| Energy contribution | -23.72 |
| Covariance contribution | 2.35 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2263005 102 + 22407834 CUCGCAUUAUCCAGCUCGAUUCAGUCGGCAAUUAAUGCCACUCCAAUUGCCCGAUUUGCCUGCACCUAC------------AUAUCCAGCCAUAUGGCUGGAUUGCGAAAGAAU .(((((.......((..(....(((((((((((...........))))).))))))..)..))......------------..((((((((....)))))))))))))...... ( -30.60) >DroSec_CAF1 23552 111 + 1 CUCGCAUUAUCCAGGCCGAUUCAGUCGGCAAUUAAUGCCACUCCAAUUGCCCGAUUUGCCUGCACCUCCGAAGGGGGUAGCAUAU--GGC-AAAUGGCUGGAUUGCGAAAGAAU .(((((..((((((((((((...)))))).....................((.(((((((((((((.((....))))).)))...--)))-)))))))))))))))))...... ( -44.10) >DroSim_CAF1 24692 112 + 1 CUCGCAUUAUCCAGCCCGAUUCAGUCGGCAAUUAAUGCCACUCCAAUUGCCCGAUUUGCCUGCACCUCCGAAUGGGGUAGCAUAU--GGCCAAAUGGCUGGAUUGCGAAAGAAU .(((((..(((((((((((((.(((.((((.....)))))))..)))))....(((((((((((((((.....))))).)))...--)).))))))))))))))))))...... ( -41.20) >DroEre_CAF1 25262 89 + 1 CUCCCAUUAUCCAGCCCGUUUCAGCCGGCAAUUAAUGGCACUCCAAUUGCCCGAUUUGCCUGUAC---------------C----------AUAUGGCUGGACUGCGAAAGAAU .........(((((((.......((.(((((.....((((.......))))....))))).))..---------------.----------....)))))))...(....)... ( -26.24) >DroYak_CAF1 31582 104 + 1 CUCACAUUAUCCAGCCCGAUUCAGGCGGCAAUUAAUGCCACUCCAAUUGCUUGAUUUGCCUGUACCUCGGAAUGAGGUAGC----------AUAUGGCUGGAUGGCGAAAGAAU ......((((((((((.....(((((((((.....))))....(((....)))....)))))((((((.....))))))..----------....))))))))))(....)... ( -37.20) >consensus CUCGCAUUAUCCAGCCCGAUUCAGUCGGCAAUUAAUGCCACUCCAAUUGCCCGAUUUGCCUGCACCUCCGAA_G_GGUAGCAUAU___GC_AUAUGGCUGGAUUGCGAAAGAAU .(((((..((((((((.......((.(((((.....((..........)).....))))).))(((((.....))))).................)))))))))))))...... (-21.37 = -23.72 + 2.35)

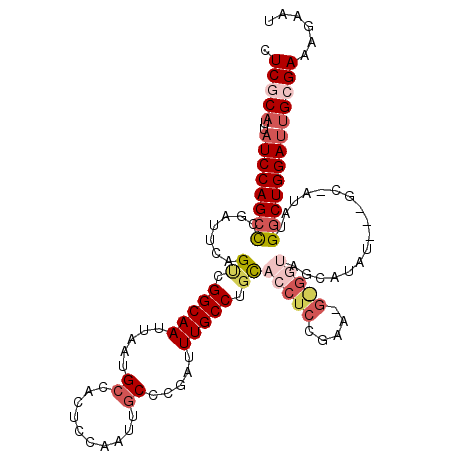
| Location | 2,263,005 – 2,263,107 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
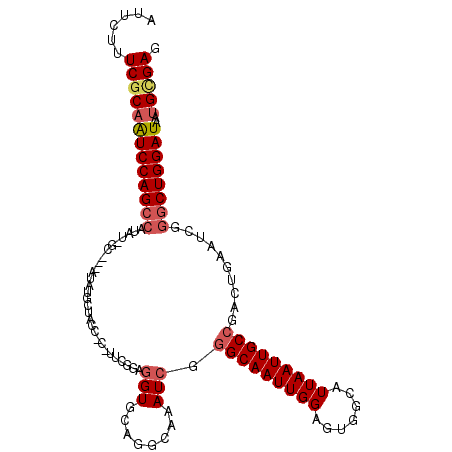
>2L_DroMel_CAF1 2263005 102 - 22407834 AUUCUUUCGCAAUCCAGCCAUAUGGCUGGAUAU------------GUAGGUGCAGGCAAAUCGGGCAAUUGGAGUGGCAUUAAUUGCCGACUGAAUCGAGCUGGAUAAUGCGAG ......(((((((((((((.....(((.(((.(------------((........))).))).)))......(((((((.....)))).))).....).)))))))..))))). ( -35.40) >DroSec_CAF1 23552 111 - 1 AUUCUUUCGCAAUCCAGCCAUUU-GCC--AUAUGCUACCCCCUUCGGAGGUGCAGGCAAAUCGGGCAAUUGGAGUGGCAUUAAUUGCCGACUGAAUCGGCCUGGAUAAUGCGAG ......(((((((((((((((((-(((--...(((.(((((....)).)))))))))))))..(((((((((.......))))))))).........)).))))))..))))). ( -42.40) >DroSim_CAF1 24692 112 - 1 AUUCUUUCGCAAUCCAGCCAUUUGGCC--AUAUGCUACCCCAUUCGGAGGUGCAGGCAAAUCGGGCAAUUGGAGUGGCAUUAAUUGCCGACUGAAUCGGGCUGGAUAAUGCGAG ......(((((((((((((.....(((--...(((.(((((....)).)))))))))......(((((((((.......)))))))))..........))))))))..))))). ( -43.30) >DroEre_CAF1 25262 89 - 1 AUUCUUUCGCAGUCCAGCCAUAU----------G---------------GUACAGGCAAAUCGGGCAAUUGGAGUGCCAUUAAUUGCCGGCUGAAACGGGCUGGAUAAUGGGAG ...((..((..((((((((....----------(---------------((...(((((....((((.......)))).....))))).)))......))))))))..))..)) ( -33.30) >DroYak_CAF1 31582 104 - 1 AUUCUUUCGCCAUCCAGCCAUAU----------GCUACCUCAUUCCGAGGUACAGGCAAAUCAAGCAAUUGGAGUGGCAUUAAUUGCCGCCUGAAUCGGGCUGGAUAAUGUGAG ......((((.((((((((....----------..((((((.....))))))(((((...((((....))))...((((.....))))))))).....))))))))...)))). ( -43.50) >consensus AUUCUUUCGCAAUCCAGCCAUAU_GC___AUAUGCUACC_C_UUCGGAGGUGCAGGCAAAUCGGGCAAUUGGAGUGGCAUUAAUUGCCGACUGAAUCGGGCUGGAUAAUGCGAG ......(((((((((((((.............................(((........))).(((((((((.......)))))))))..........))))))))..))))). (-23.70 = -24.62 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:44 2006