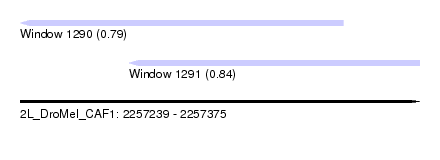
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,257,239 – 2,257,375 |
| Length | 136 |
| Max. P | 0.838665 |

| Location | 2,257,239 – 2,257,349 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -40.11 |
| Consensus MFE | -33.14 |
| Energy contribution | -33.56 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
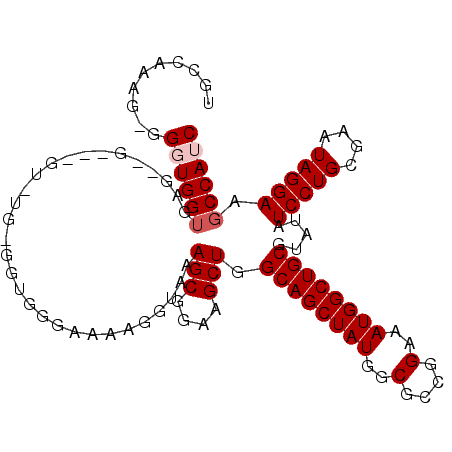
>2L_DroMel_CAF1 2257239 110 - 22407834 ---UAAAAGGUAA-AGCGGAAGCUGGCAGCUAUGGCGC---CGGAAAUGGCUGCAUAUAUCCUGCGAAUAGGAAGCCAUCGCUAUCGCCAACGCUGGCAAUAGUUUUGCGGUGCUU-U ---...((((((.-.((((((..((.((((..(((((.---.((..((((((.......(((((....)))))))))))..))..)))))..)))).))....))))))..)))))-) ( -40.11) >DroSec_CAF1 18008 110 - 1 ---GAAAAGGUAA-AGCGGAAGCUGGCAGCUAUGGCGC---CGGAAAUGGCUGCGUAUAUCCUGCGAAUAGGAAGCCAUCGCUAUCGCCAUCGUUGGCAAUAGUUUUGCGGUGCUU-U ---...((((((.-.((((((..((.((((.((((((.---.((..((((((.......(((((....)))))))))))..))..)))))).)))).))....))))))..)))))-) ( -40.41) >DroSim_CAF1 19046 110 - 1 ---GAAAAGGUAA-AGCGGAAGCUGGCAGCUAUGGCGC---CGGAAAUGGCUGCGUAUAUCCUGCGAAUAGGAAGCCAUCGCUAUCGCCAUCGUUGGCAAUAGUUUUGCGGUGCUU-U ---...((((((.-.((((((..((.((((.((((((.---.((..((((((.......(((((....)))))))))))..))..)))))).)))).))....))))))..)))))-) ( -40.41) >DroEre_CAF1 19557 110 - 1 ---UAGAAGGUGA-AGCGGAAGCUGGCAGCUAUGGCGC---CGGAAAUGGCUGCGUAUAUCCUGCGAAUAGGAAGCCAUCGCUAUCGCCGUCGCUGGCAAUAGUUUUGCGGUGCUU-U ---..((.(((((-(((((..(((.((((((((..(..---..)..)))))))).....(((((....))))))))..))))).))))).))(((.((((.....)))).).))..-. ( -42.00) >DroYak_CAF1 25618 111 - 1 ---AGGAAGGUAA-AGCGGAAGCUGGCAGCUAUGGCGC---CGGAAAUGGCUGCGUAUAUCCUGCGAAUAGGAAGCCAACGCUAUCGCCAUCGCUGGCAAUAGUUUUGCGGUGCUUUU ---..(((((((.-.((((((..((.((((.(((((((---((....)))).((((...(((((....))))).....))))....))))).)))).))....))))))..))))))) ( -42.30) >DroAna_CAF1 21038 116 - 1 ACUGAAAAGGUAAAAACGGAAACUGCCAGCUCCAGCUCCAACGGAAAUGGCUGCGUAUAUCCUG-GAAUAGGAAGCCAAAGCUAUCGCCAUUGCUGGCAAUAGUUUUGUGGUGCUU-U ......((((((...((((((..((((((((((.........)))((((((........(((((-...)))))(((....)))...)))))))))))))....))))))..)))))-) ( -35.40) >consensus ___GAAAAGGUAA_AGCGGAAGCUGGCAGCUAUGGCGC___CGGAAAUGGCUGCGUAUAUCCUGCGAAUAGGAAGCCAUCGCUAUCGCCAUCGCUGGCAAUAGUUUUGCGGUGCUU_U .......(((((...((((((..((.((((.((((((.....((..((((((.......(((((....)))))))))))..))..)))))).)))).))....))))))..))))).. (-33.14 = -33.56 + 0.42)


| Location | 2,257,276 – 2,257,375 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -26.54 |
| Energy contribution | -27.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2257276 99 - 22407834 UGUCAAAGGGGGUGGUGC----------UGUGGUGGUAAAAGGUAAAGCGGAAGCUGGCAGCUAUGGCGCCGGAAAUGGCUGCAUAUAUCCUGCGAAUAGGAAGCCAUC ............((((((----------((((((.((.........(((....))).)).))))))))))))...((((((.......(((((....))))))))))). ( -32.71) >DroSec_CAF1 18045 108 - 1 UGCCAAAG-GGGUGGUGAGGAGGAGGUGUGCGGUGGGAAAAGGUAAAGCGGAAGCUGGCAGCUAUGGCGCCGGAAAUGGCUGCGUAUAUCCUGCGAAUAGGAAGCCAUC ........-.((((((.....(.((((((((...............(((....))).((((((((..(....)..))))))))))))).))).).........)))))) ( -34.34) >DroSim_CAF1 19083 108 - 1 UGCCAAGG-GGGUGGUGAGGAGGAGGUGUGUGGUGGGAAAAGGUAAAGCGGAAGCUGGCAGCUAUGGCGCCGGAAAUGGCUGCGUAUAUCCUGCGAAUAGGAAGCCAUC .((((...-...)))).....((...(.(((.(..(((........(((....))).((((((((..(....)..)))))))).....)))..)..))).)...))... ( -34.00) >DroEre_CAF1 19594 92 - 1 UGCCCCGGGGGUUGG-GU----------------GGUAGAAGGUGAAGCGGAAGCUGGCAGCUAUGGCGCCGGAAAUGGCUGCGUAUAUCCUGCGAAUAGGAAGCCAUC .((((...))))..(-((----------------(((.........(((....))).((((((((..(....)..)))))))).....(((((....))))).)))))) ( -32.20) >DroYak_CAF1 25656 102 - 1 UGCCCAAGUGGGUGGUGUGUUG---GU----GUAGGAGGAAGGUAAAGCGGAAGCUGGCAGCUAUGGCGCCGGAAAUGGCUGCGUAUAUCCUGCGAAUAGGAAGCCAAC .((((....))))(((......---.(----((((((.........(((....))).((((((((..(....)..)))))))).....)))))))........)))... ( -38.06) >consensus UGCCAAAG_GGGUGGUGAG__G___GU_UG_GGUGGGAAAAGGUAAAGCGGAAGCUGGCAGCUAUGGCGCCGGAAAUGGCUGCGUAUAUCCUGCGAAUAGGAAGCCAUC ..........((((((..............................(((....))).((((((((..(....)..)))))))).....(((((....))))).)))))) (-26.54 = -27.14 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:40 2006