| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,248,795 – 2,248,925 |
| Length | 130 |
| Max. P | 0.981951 |

| Location | 2,248,795 – 2,248,885 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -19.90 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
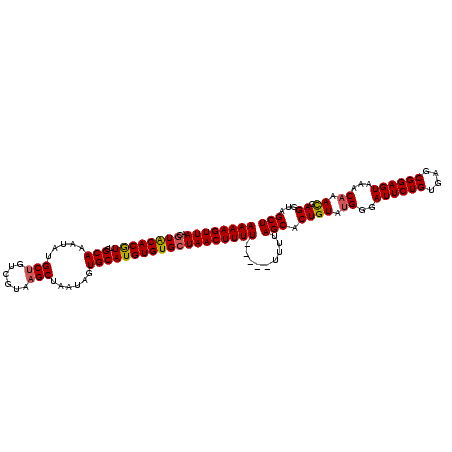
>2L_DroMel_CAF1 2248795 90 + 22407834 UGGUGGUCGAAAAAGUUAACGUACACGUUUGCAAAUAUGCUGUCGUAAGCUAAUAGUGCAUGUGUGCUAACUUUUUUUUUUUUUAGCACU .((((.(.((((((((((..(((((((..((((.....(((......)))......))))))))))))))))))))).......).)))) ( -23.20) >DroSec_CAF1 9021 85 + 1 UGGUGGUCGAAAAAGUUAACGUACACAUUCGCAAAUAUGCUGUCGUAAGCUAAUAGUGCAUGUGUGAUAACUUUU-----UUUUAGCACU .((((.(.((((((((((...(((((((.(((......(((......))).....))).))))))).))))))))-----))..).)))) ( -24.40) >DroSim_CAF1 9284 85 + 1 UGGUGGUCGAAAAAGUUAACGUACACAUUCGCAAAUAUGCUGUUGUAAGCUAAUAGUGCAUGUGUGCUAACUUUU-----UUUUAGCACU .((((.(.((((((((((..((((((((.(((......(((......))).....))).))))))))))))))))-----))..).)))) ( -26.60) >DroEre_CAF1 9077 85 + 1 UGGUGGUCGAAAAAGUUAACGUGCACGUUCGCAAAUAUGCUGUCGUAAGCUAAUAGUGCAUGUGUGCUAACUUUU-----UUGUAGCACU .((((.((((((((((((..(..(((((.(((......(((......))).....))).)))))..)))))))))-----))).).)))) ( -29.10) >DroYak_CAF1 9367 85 + 1 UGGUGGUCGAAAAAGUUAACGUACACGUUCGCAAAUAUGCUGUCGUGCGCUAAUAGUGCAUGUGUGCUAACUUUU-----UUUCAGCACU .((((.(.((((((((((..((((((....(((....)))....((((((.....))))))))))))))))))))-----))..).)))) ( -28.00) >consensus UGGUGGUCGAAAAAGUUAACGUACACGUUCGCAAAUAUGCUGUCGUAAGCUAAUAGUGCAUGUGUGCUAACUUUU_____UUUUAGCACU .((((.(.((((((((((..((((((((..(((.....(((......)))......))))))))))))))))))......))).).)))) (-19.90 = -20.10 + 0.20)


| Location | 2,248,795 – 2,248,885 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -14.94 |
| Energy contribution | -14.58 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2248795 90 - 22407834 AGUGCUAAAAAAAAAAAAAGUUAGCACACAUGCACUAUUAGCUUACGACAGCAUAUUUGCAAACGUGUACGUUAACUUUUUCGACCACCA ..............(((((((((((((((.((((......(((......))).....))))...))))..)))))))))))......... ( -16.90) >DroSec_CAF1 9021 85 - 1 AGUGCUAAAA-----AAAAGUUAUCACACAUGCACUAUUAGCUUACGACAGCAUAUUUGCGAAUGUGUACGUUAACUUUUUCGACCACCA .(((.....(-----((((((((..((((((((.......))........(((....)))..))))))....)))))))))....))).. ( -16.80) >DroSim_CAF1 9284 85 - 1 AGUGCUAAAA-----AAAAGUUAGCACACAUGCACUAUUAGCUUACAACAGCAUAUUUGCGAAUGUGUACGUUAACUUUUUCGACCACCA .(((.....(-----((((((((((((((((((.......))........(((....)))..))))))..)))))))))))....))).. ( -20.30) >DroEre_CAF1 9077 85 - 1 AGUGCUACAA-----AAAAGUUAGCACACAUGCACUAUUAGCUUACGACAGCAUAUUUGCGAACGUGCACGUUAACUUUUUCGACCACCA .(((...(.(-----((((((((((......((.......)).((((...(((....)))...))))...))))))))))).)..))).. ( -18.60) >DroYak_CAF1 9367 85 - 1 AGUGCUGAAA-----AAAAGUUAGCACACAUGCACUAUUAGCGCACGACAGCAUAUUUGCGAACGUGUACGUUAACUUUUUCGACCACCA .(((.....(-----((((((((((......((.......))(((((...(((....)))...)))))..)))))))))))....))).. ( -22.80) >consensus AGUGCUAAAA_____AAAAGUUAGCACACAUGCACUAUUAGCUUACGACAGCAUAUUUGCGAACGUGUACGUUAACUUUUUCGACCACCA ...............((((((((((......((.......)).((((...(((....)))...))))...)))))))))).......... (-14.94 = -14.58 + -0.36)

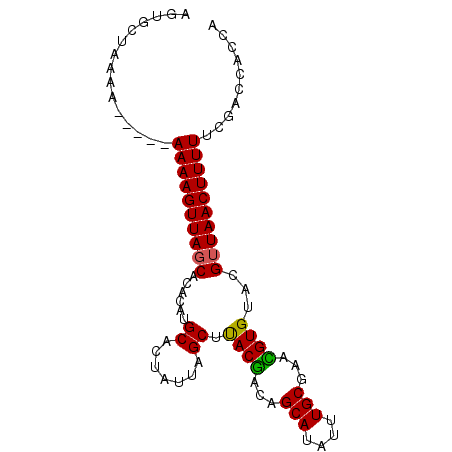
| Location | 2,248,805 – 2,248,925 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.79 |
| Mean single sequence MFE | -32.56 |
| Consensus MFE | -27.56 |
| Energy contribution | -27.90 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2248805 120 + 22407834 AAAAGUUAACGUACACGUUUGCAAAUAUGCUGUCGUAAGCUAAUAGUGCAUGUGUGCUAACUUUUUUUUUUUUUAGCACUGUUUGGGAUUCUGUGAGCGGAGUAAACAAACGAGGUAGCU ((((((((..(((((((..((((.....(((......)))......))))))))))))))))))).........(((((((((((..((((((....))))))...)))))..))).))) ( -34.50) >DroSec_CAF1 9031 115 + 1 AAAAGUUAACGUACACAUUCGCAAAUAUGCUGUCGUAAGCUAAUAGUGCAUGUGUGAUAACUUUU-----UUUUAGCACUGUGUGGGAUUCUGUGAGCGGAGUAAACAAACGAGGUAGCU ((((((((...(((((((.(((......(((......))).....))).))))))).))))))))-----....((((((.(((...((((((....))))))..))).....))).))) ( -30.30) >DroSim_CAF1 9294 115 + 1 AAAAGUUAACGUACACAUUCGCAAAUAUGCUGUUGUAAGCUAAUAGUGCAUGUGUGCUAACUUUU-----UUUUAGCACUGUGUGGGAUUCUGUGAGCGGAGUAAACAAACGAGAUAGCU ((((((((..((((((((.(((......(((......))).....))).))))))))))))))))-----....(((.((((.((..((((((....))))))...)).)).))...))) ( -29.80) >DroEre_CAF1 9087 115 + 1 AAAAGUUAACGUGCACGUUCGCAAAUAUGCUGUCGUAAGCUAAUAGUGCAUGUGUGCUAACUUUU-----UUGUAGCACUGUACGGCAUUCUGUGAGCGGAGUAAACAAACGAGUUGGCU ...(((((((.(((.((((((((...((((((((....((.......))....((((((.(....-----..))))))).).)))))))..))))))))..))).........))))))) ( -35.30) >DroYak_CAF1 9377 115 + 1 AAAAGUUAACGUACACGUUCGCAAAUAUGCUGUCGUGCGCUAAUAGUGCAUGUGUGCUAACUUUU-----UUUCAGCACUGUAUGGGAUUCUGUGAACGGAGUAAACAAAUGAGUUGGCU ...(((((((.(((.((((((((..(((((...(((((((.....))))))).(((((.......-----....))))).)))))......))))))))..))).........))))))) ( -32.90) >consensus AAAAGUUAACGUACACGUUCGCAAAUAUGCUGUCGUAAGCUAAUAGUGCAUGUGUGCUAACUUUU_____UUUUAGCACUGUAUGGGAUUCUGUGAGCGGAGUAAACAAACGAGGUAGCU ((((((((..((((((((..(((.....(((......)))......))))))))))))))))))).........(((.((((.((..((((((....))))))...)).)).))...))) (-27.56 = -27.90 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:36 2006