
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 22,227,890 – 22,228,170 |
| Length | 280 |
| Max. P | 0.889390 |

| Location | 22,227,890 – 22,228,010 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -35.79 |
| Energy contribution | -35.47 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22227890 120 + 22407834 ACUACAAUGUAAAGUGCUUGUCUAUGGGCAUGCUGACACCUGUAGAGACCUCUGUUAUCUGGCGAGGUCCUCUUGUAAUGUCAGCAAUACAACGACUGCUAAAAGGUACUGAUUGGGGGC .(..((((....((((((((((....))).((((((((.....(((((((((.(((....)))))))).)))).....)))))))).................))))))).))))..).. ( -37.70) >DroSim_CAF1 27648 120 + 1 ACUACAAUGUAAAGUGCUUGUCUAUGGGCAUGCUGACACCUGUAGAGGCCUCUGUUAUCUGGCGAGGUCCUCUUGUAAUGUCAGCAAUACAACGACUGUUAAAAGGUACUGAUUGGGGAC .(..((((....((((((((((....))).((((((((.....(((((((((.(((....))))))).))))).....)))))))).................))))))).))))..).. ( -39.60) >DroYak_CAF1 223718 120 + 1 ACUACAAUGUAAAGUGCUUGUCUAUGGGCAUGUUGACACCUGUUGAGACCUCUGUUAUAUGGCGAGGUCCUCUUGUAAUGUCAGCAAUACGACGGCUGCUAAAAGGUGCUGAUUGGGGAC .(..(((((((..((((((......))))))(((((((......((((((((.(((....)))))))).)))......)))))))..)))..((((.(((....)))))))))))..).. ( -35.90) >consensus ACUACAAUGUAAAGUGCUUGUCUAUGGGCAUGCUGACACCUGUAGAGACCUCUGUUAUCUGGCGAGGUCCUCUUGUAAUGUCAGCAAUACAACGACUGCUAAAAGGUACUGAUUGGGGAC .(..((((....((((((((((....))).((((((((.....(((((((((.(((....)))))))).)))).....)))))))).................))))))).))))..).. (-35.79 = -35.47 + -0.33)

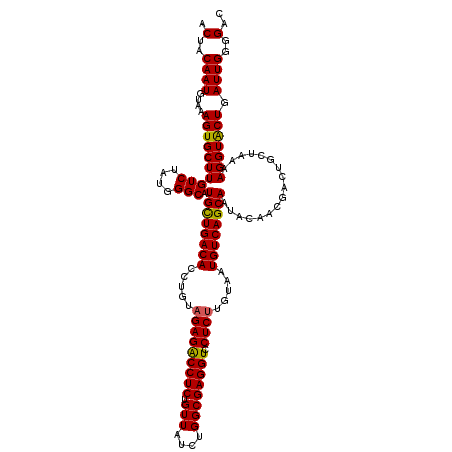
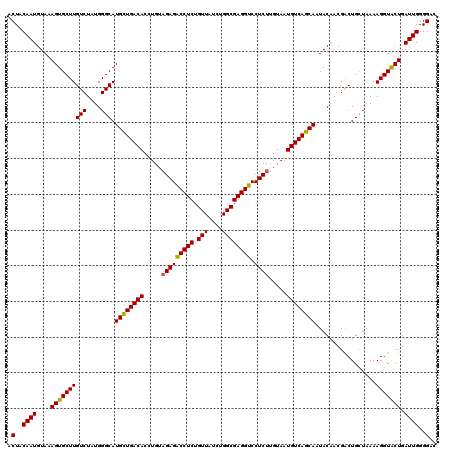
| Location | 22,228,010 – 22,228,130 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -34.64 |
| Energy contribution | -34.20 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22228010 120 - 22407834 GAACAGCUGCAGUAUGAGGUGUUGUUACCAGGAUAACCCCUGUGAUAGGUGCAUGCUGGGAUAGUGAGAGAUGUACAUCUCCAGUGCCCGGUGGAGUAUCUAUAACUAGGACAUCUAGAA .((((((.((........)))))))).(((((......)))).)((((((((.(((((((.((.((.(((((....))))))).)))))))))..))))))))..((((.....)))).. ( -39.50) >DroSim_CAF1 27768 120 - 1 GAACUGCUGCAGUAUGAGGUGUUGUUACAAGGAUAACCCCAGUGAUGGGUGUAUGCUGGGAUAGUGAGAGAUGCACAUCUCCAGUGCCCGGUGGAGUAUCUAUAACCAGGACAUCUAAAA ..((((...))))...(((((((((((...(((((((((((....)))).))..((((((.((.((.(((((....))))))).))))))))....))))).))))...))))))).... ( -38.00) >DroYak_CAF1 223838 120 - 1 GAACAGCUGCAGUAUGAGGUGUUGUUACAAGGAUAACACCUGUGAUAGGUGCAUGCUGGGAUAGUGAGAGAUGUACAUCUCCAGUGCCCGGUGGAGUAUCUAUAACCAAUACAUCUAAAA ...........((((.((((((((((.....))))))))))((.((((((((.(((((((.((.((.(((((....))))))).)))))))))..)))))))).))..))))........ ( -42.50) >consensus GAACAGCUGCAGUAUGAGGUGUUGUUACAAGGAUAACCCCUGUGAUAGGUGCAUGCUGGGAUAGUGAGAGAUGUACAUCUCCAGUGCCCGGUGGAGUAUCUAUAACCAGGACAUCUAAAA ..........((.(((.((.((((((.....)))))).)).((.((((((((.(((((((.((.((.(((((....))))))).)))))))))..)))))))).)).....))))).... (-34.64 = -34.20 + -0.44)

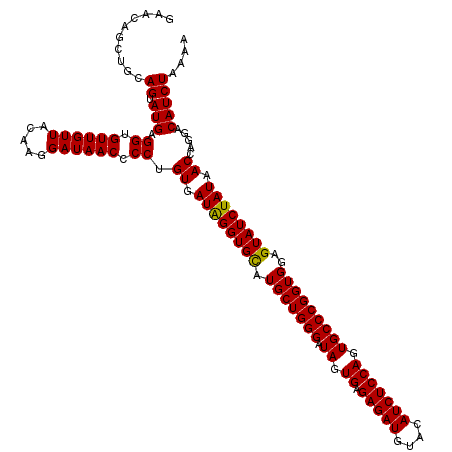
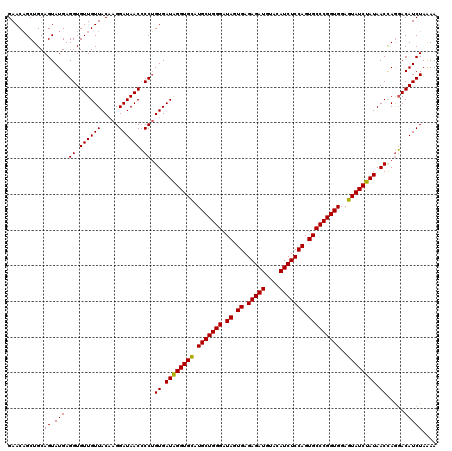
| Location | 22,228,050 – 22,228,170 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -26.12 |
| Energy contribution | -27.57 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22228050 120 + 22407834 AGAUGUACAUCUCUCACUAUCCCAGCAUGCACCUAUCACAGGGGUUAUCCUGGUAACAACACCUCAUACUGCAGCUGUUCAGGUGACCUUGAAAGGUGCAAGCAUGUACGAAAAGUUAAA ...(((((((..............((.(((((((.(((.(((.(((((....)))))..(((((...((.......))..))))).)))))).))))))).))))))))).......... ( -35.33) >DroSim_CAF1 27808 120 + 1 AGAUGUGCAUCUCUCACUAUCCCAGCAUACACCCAUCACUGGGGUUAUCCUUGUAACAACACCUCAUACUGCAGCAGUUCAGGUGACCUUGAAAGGUGCAAGGAUGUACGAAAAGCUAAA ...(((((..........(((((((.............))))))).((((((((.....(((((...((((...))))..)))))((((....))))))))))))))))).......... ( -35.12) >DroYak_CAF1 223878 120 + 1 AGAUGUACAUCUCUCACUAUCCCAGCAUGCACCUAUCACAGGUGUUAUCCUUGUAACAACACCUCAUACUGCAGCUGUUCAGGUGACCUUAAAAGGUGCCAGGAUGUACGAAAAGUUAAA ...(((((((((..........((((.((((........(((((((...........))))))).....))))))))....(((.((((....))))))).))))))))).......... ( -33.12) >consensus AGAUGUACAUCUCUCACUAUCCCAGCAUGCACCUAUCACAGGGGUUAUCCUUGUAACAACACCUCAUACUGCAGCUGUUCAGGUGACCUUGAAAGGUGCAAGGAUGUACGAAAAGUUAAA ...(((((((((....((.....))...((((((.(((.(((.(((((....)))))..(((((...((.......))..))))).)))))).))))))..))))))))).......... (-26.12 = -27.57 + 1.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:44 2006