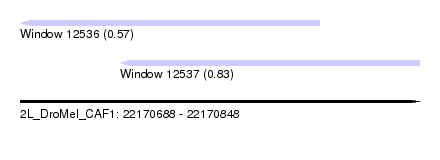
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 22,170,688 – 22,170,848 |
| Length | 160 |
| Max. P | 0.832467 |

| Location | 22,170,688 – 22,170,808 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -22.19 |
| Energy contribution | -21.94 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22170688 120 - 22407834 GCAACUUUUCGUUGUACAAAGGCAGAGAAGACAUAAGUACAAAGAACGCCGUGGUACCCUGCGAGUGUGCAACAAAAUGCAAAGAGCUGGACUGACUCUUCGCCAGCGCAUAGUCUAGCA (((..((((.((((((((...((((.(....)....((((...(.....)...)))).))))...))))))))))))))).....(((((((((......((....))..))))))))). ( -32.80) >DroAna_CAF1 1510 120 - 1 GCACCUUCUCGUUGUACCAGGGUAGCGAAGACAUCAACACAAAGAAUGUAGUUGUUCCCUGCGAAUGUGCUACAAAAUGAAGCGAGUUCGAUUUGCUUUCCACAAGCGCAUAAUCGAGCA ((...............(((((.(((((..(((((........).))))..))))))))))((((((((((.......(((((((((...))))))))).....)))))))..))).)). ( -33.70) >DroEre_CAF1 43824 120 - 1 GCAACUUUUCGUUGUACAAAGGCAGAGAAGACAUAAGAACGAAAAAUGUCGUGGUGCCCUGCGAGUGUGCAACGAAAUGCAAAGAGCUGGACUGACUCUCCUCCAGCGCAUAGUCCAGCA (((...((((((((((((...((((.(..(((((...........)))))......).))))...))))))))))))))).....(((((((((.(.((.....)).)..))))))))). ( -42.10) >DroYak_CAF1 38361 120 - 1 GCAACUUCUCGUUGUACAAAGGCAGAGAAGACAUAAGUACGAAAAACGUCGUGGUGCCUUGCGAAUGUGCAACAAAAUGCAAAGAGUUUGACUGACUCUCCGCUAGCGCAUAGUCCAGCA ((...((((((((.......))).)))))(((.....(((((......)))))((((((.(((....((((......)))).((((((.....)))))).))).)).)))).)))..)). ( -32.20) >consensus GCAACUUCUCGUUGUACAAAGGCAGAGAAGACAUAAGUACAAAAAACGUCGUGGUGCCCUGCGAAUGUGCAACAAAAUGCAAAGAGCUGGACUGACUCUCCGCCAGCGCAUAGUCCAGCA ..........((((((((...((((.(....(((................)))...).))))...))))))))............(((((((((...((.....))....))))))))). (-22.19 = -21.94 + -0.25)



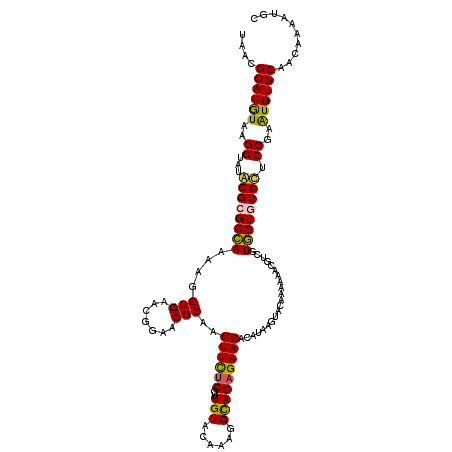
| Location | 22,170,728 – 22,170,848 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -26.02 |
| Energy contribution | -25.53 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22170728 120 - 22407834 UCACGCAUGUAAGCUAUGGGCGCCAAUAGGUGCACGGAACGCAACUUUUCGUUGUACAAAGGCAGAGAAGACAUAAGUACAAAGAACGCCGUGGUACCCUGCGAGUGUGCAACAAAAUGC ....(((..(..((...(((..(((...((((..(.(.((.....(((((.((((......)))).))))).....)).)...)..)))).)))..))).))..)..))).......... ( -32.40) >DroAna_CAF1 1550 120 - 1 UUUCGCAUAUACGCAAUGGGCGCAAGAAGGUGAACAGAUCGCACCUUCUCGUUGUACCAGGGUAGCGAAGACAUCAACACAAAGAAUGUAGUUGUUCCCUGCGAAUGUGCUACAAAAUGA ....((((((.((((.(((((((.((((((((.........)))))))).).))).)))(((.(((((..(((((........).))))..)))))))))))).)))))).......... ( -40.10) >DroEre_CAF1 43864 120 - 1 UAACGCAUGUAAGCUAUAGGCGCCAAUAAGUGAACGGAACGCAACUUUUCGUUGUACAAAGGCAGAGAAGACAUAAGAACGAAAAAUGUCGUGGUGCCCUGCGAGUGUGCAACGAAAUGC ....(((..(..((....(((((((....((((((((((.......))))))).)))............(((((...........))))).)))))))..))..)..))).......... ( -31.30) >DroYak_CAF1 38401 120 - 1 UAUCGCAUGUAAGCUAUAGGCGCCAAAAGGUGAACGGAACGCAACUUCUCGUUGUACAAAGGCAGAGAAGACAUAAGUACGAAAAACGUCGUGGUGCCUUGCGAAUGUGCAACAAAAUGC ....((((((..((...((((((((..(.((...((..((....(((((((((.......))).))))))......)).))....)).)..)))))))).))..)))))).......... ( -35.10) >consensus UAACGCAUGUAAGCUAUAGGCGCCAAAAGGUGAACGGAACGCAACUUCUCGUUGUACAAAGGCAGAGAAGACAUAAGUACAAAAAACGUCGUGGUGCCCUGCGAAUGUGCAACAAAAUGC ....((((((..((...((((((((....(((.......)))..((((((..(((......))))))))).....................)))))))).))..)))))).......... (-26.02 = -25.53 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:41 2006