
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 22,153,929 – 22,154,037 |
| Length | 108 |
| Max. P | 0.748793 |

| Location | 22,153,929 – 22,154,037 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -21.81 |
| Energy contribution | -21.00 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
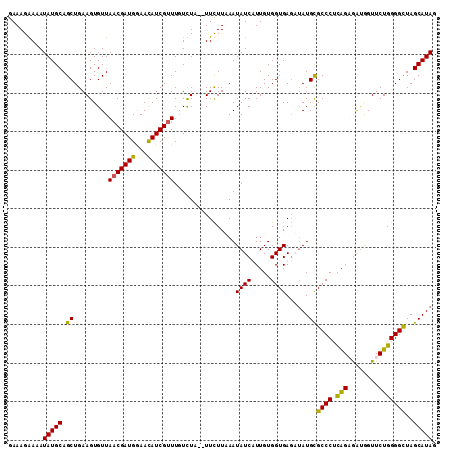
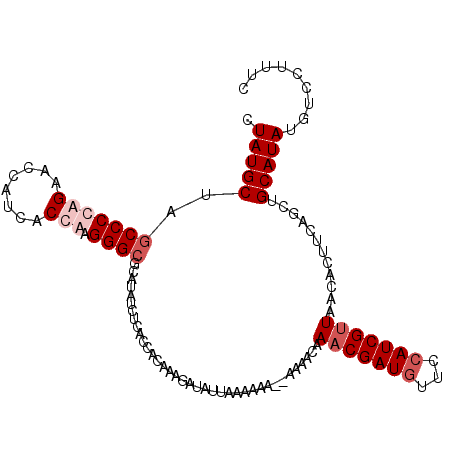
>2L_DroMel_CAF1 22153929 108 + 22407834 GAAAGGACAUAUGCAGCUGAAGUGUUAACGAUGGAACAUCGUUU-UCUA--UUCUUUAAUAUCAUUGUGGUGAUAUAUGCGCCCUUAGAGAUGGUUCUGGGGCUAGCAUAG .........(((((.(((((((.(..(((((((...))))))).-.)..--..)))))(((((((....)))))))..))(((((.((((....)))))))))..))))). ( -32.10) >DroSim_CAF1 25922 109 + 1 GAAAGGACAUAUGCAGCUGAAGUGUUAACGAUGGAAUAUCGUUUGUCUA--UUCUUUAAUAUCAUUGUGGUGAGAUAUGCGCCCGCAGAGGUGGUUCUGGGGUUAGCAUAG (((.(((((.....(((......)))(((((((...)))))))))))).--))).......((((....))))..(((((((((.(((((....)))))))))..))))). ( -28.30) >DroEre_CAF1 30938 110 + 1 GAAAGAAAAUAUGCAGCUGAAGUGUUACCGAUGAAACAUCGUUUGUUUUGUUUUUUAAAUAUCUUUGUGGUGAG-AAUGUGCCCUUGGUAAUGGUUCUGGGGCUAGCAUAG .........((((((((((((.(((((((((..(((((.....)))))..)).........((((......)))-)..........))))))).)))...)))).))))). ( -27.40) >DroYak_CAF1 32602 109 + 1 GAAAGAAAAUAUGCAGCUGAAGUGUUAACGAUGGAACAUCGUUUAUUUU--UUUUUAAAUAUCUUUGUGGUGAGUUAUGCGCCCUCGGUGACAGUUCUGGGGCUAGCAUAG .........((((((((((((.(((((.(((.((..(((..((((((..--.................))))))..)))..)).))).))))).)))...)))).))))). ( -27.61) >consensus GAAAGAAAAUAUGCAGCUGAAGUGUUAACGAUGGAACAUCGUUUGUCUA__UUCUUAAAUAUCAUUGUGGUGAGAUAUGCGCCCUCAGAGAUGGUUCUGGGGCUAGCAUAG .........(((((.((.........(((((((...)))))))................((((.....))))......))((((.(((........)))))))..))))). (-21.81 = -21.00 + -0.81)


| Location | 22,153,929 – 22,154,037 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -20.45 |
| Consensus MFE | -13.30 |
| Energy contribution | -15.05 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
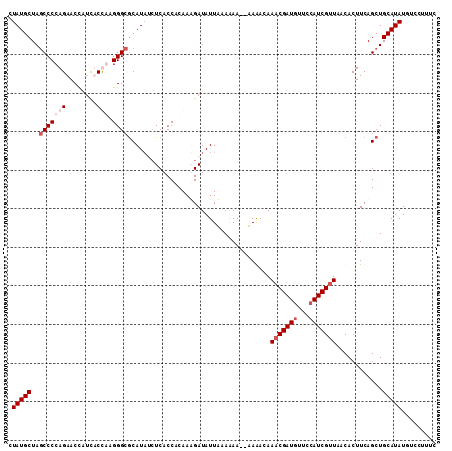
>2L_DroMel_CAF1 22153929 108 - 22407834 CUAUGCUAGCCCCAGAACCAUCUCUAAGGGCGCAUAUAUCACCACAAUGAUAUUAAAGAA--UAGA-AAACGAUGUUCCAUCGUUAACACUUCAGCUGCAUAUGUCCUUUC .(((((..((((.(((......)))..))))((..((((((......))))))....(((--....-.(((((((...))))))).....))).)).)))))......... ( -24.50) >DroSim_CAF1 25922 109 - 1 CUAUGCUAACCCCAGAACCACCUCUGCGGGCGCAUAUCUCACCACAAUGAUAUUAAAGAA--UAGACAAACGAUAUUCCAUCGUUAACACUUCAGCUGCAUAUGUCCUUUC .(((((...(((((((......)))).))).(((((((..........)))))....(((--......((((((.....)))))).....))).)).)))))......... ( -21.30) >DroEre_CAF1 30938 110 - 1 CUAUGCUAGCCCCAGAACCAUUACCAAGGGCACAUU-CUCACCACAAAGAUAUUUAAAAAACAAAACAAACGAUGUUUCAUCGGUAACACUUCAGCUGCAUAUUUUCUUUC .(((((.(((....(((...(((((..((.......-....))............................((((...)))))))))...))).))))))))......... ( -16.50) >DroYak_CAF1 32602 109 - 1 CUAUGCUAGCCCCAGAACUGUCACCGAGGGCGCAUAACUCACCACAAAGAUAUUUAAAAA--AAAAUAAACGAUGUUCCAUCGUUAACACUUCAGCUGCAUAUUUUCUUUC .(((((..((((..(.........)..))))((.................(((((.....--.)))))(((((((...))))))).........)).)))))......... ( -19.50) >consensus CUAUGCUAGCCCCAGAACCAUCACCAAGGGCGCAUAUCUCACCACAAAGAUAUUAAAAAA__AAAACAAACGAUGUUCCAUCGUUAACACUUCAGCUGCAUAUGUCCUUUC .(((((..(((((((........))).)))).....................................(((((((...)))))))............)))))......... (-13.30 = -15.05 + 1.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:38 2006