
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 22,146,229 – 22,146,344 |
| Length | 115 |
| Max. P | 0.711088 |

| Location | 22,146,229 – 22,146,344 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.72 |
| Mean single sequence MFE | -42.97 |
| Consensus MFE | -28.90 |
| Energy contribution | -29.73 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
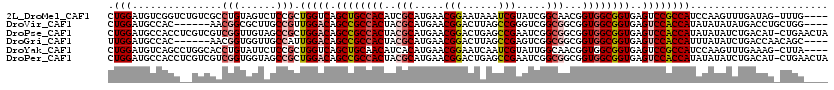
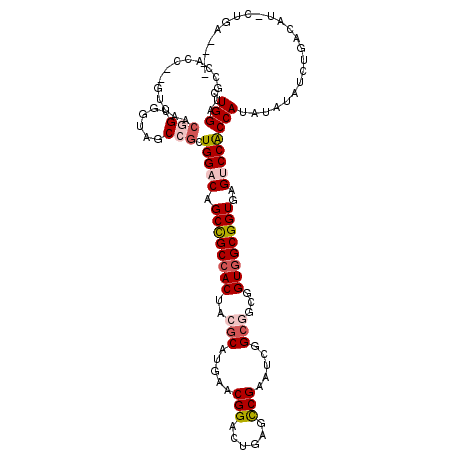
>2L_DroMel_CAF1 22146229 115 + 22407834 CUGGAUGUCGGUCUGUCGCCUGUAGUCUCCGCUGGUCAGCUGCCACAUCGCAUGAACGGAAUAAAUCGUAUCGGCAACGGUGGCGGUGAGUCCGCCAUCCAAGUUUGAUAG-UUUG---- .((((((.(((.(..(((((..........(((....))).(((((...((.(((((((......)))).)))))....))))))))))).))).))))))..........-....---- ( -37.40) >DroVir_CAF1 31446 110 + 1 CUGGAUGCCAC------AACGGCGCUUGCCGUUGGACAGCCGCCACUACGCAUGAACGGACUUAGCCGGGUCGGCGGCGGUGGCGGUGAGUCCACCAUAUAUAUAUGACCUGCUGG---- .(((.......------((((((....))))))((((.(((((((((.(((.(((.(((......)))..))))))..)))))))))..)))).)))...........((....))---- ( -47.00) >DroPse_CAF1 50059 119 + 1 CUGGAUGCCACCUCGUCGUCGGUUGUAGCCGCUGGACAGCCGCCACUACGCAUGAACGGACUGAGCCGAAUCGGCGGCGGUGGCGGUGAGUCCACCAUAUAUAUCUGACAU-CUGAACUA ...((((......))))(((((.((((...(.(((((.(((((((((.(((.(((.(((......)))..))))))..)))))))))..))))).)...)))).)))))..-........ ( -46.50) >DroGri_CAF1 33864 110 + 1 UUGGAUGCCAC------AACGGUGGUUGCCAUUGGACAGCCGCCACUACGCAUGAACGGACUUAGCCGAGUCGGCGGCGGUGGCGGUGAGUCCACCAUUUAUAUCUGACCAACAGC---- .(((..(((((------....)))))..))).(((((.(((((((((.(((.(((.(((......)))..))))))..)))))))))..)))))..........(((.....))).---- ( -45.50) >DroYak_CAF1 22878 115 + 1 CUGGAUGUCAGCCUGGCACCUGUAUUCUCCGCUGGUCAGCUGCAACAUCACAUGAACGGAAUCAAUCGUAUUGGCAACGGUGGCGGUGAGUCCGCCAUCCAAGUUUGAAAG-CUUA---- .(((((((((((.((((....((.......))..))))))))..))))).)).....(((.........((((....))))(((((.....))))).)))(((((....))-))).---- ( -32.00) >DroPer_CAF1 49960 119 + 1 CUGGAUGCCACCUCGUCGUCGGUGGUAGCCGCUGGACAGCCGCCACUACGCAUGAACGGACUGAGCCGAAUCGGCGGCGGUGGCGGUGAGUCCACCAUAUAUAUCUGACAU-CUGAACUA ..((.(((((((........))))))).))(.(((((.(((((((((.(((.(((.(((......)))..))))))..)))))))))..))))).)...............-........ ( -49.40) >consensus CUGGAUGCCACC__GUCAACGGUGGUAGCCGCUGGACAGCCGCCACUACGCAUGAACGGACUGAGCCGAAUCGGCGGCGGUGGCGGUGAGUCCACCAUAUAUAUCUGACAU_CUGA____ .(((...............(((......))).(((((.((((((((..(((.....(((......))).....)))...))))))))..))))))))....................... (-28.90 = -29.73 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:35 2006