
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 22,133,959 – 22,134,107 |
| Length | 148 |
| Max. P | 0.968972 |

| Location | 22,133,959 – 22,134,051 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -33.38 |
| Energy contribution | -33.22 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22133959 92 - 22407834 GUUAGGAUGCCACUGCGUGGUGGCAUCUUGGGCAGUGGCGGCUUGCCAGUCACCUUUUGCAGAGUUGAGGUCUUGCCCAGCUGUUCCUGUUG ..((((((((((((....))))))))))))(((((..((((((.((.((..(((((..........))))))).))..))))))..))))). ( -39.50) >DroSec_CAF1 6147 91 - 1 GUUAGGAUGC-ACUUCGUUGUGGCAUCUUGGGCAGUGGCGGCUUGCCAGUCACCCUUUGCAGAGUUGAGGUCUUGCCCAGCUGUUCCUGUUG ..((((((((-((......)).))))))))(((((..((((((.((.((....((((.((...)).)))).)).))..))))))..))))). ( -31.10) >DroSim_CAF1 6530 92 - 1 GUUAGGAUGCCACUUCGUGGUGGCAUCUUGGGCAGUGGCGGCUUGCCAGUCACCCUUUGCAGAGUUGAGGUCUUGCCCAGCUGUUCCUGUUG ..((((((((((((....))))))))))))(((((..((((((.((.((....((((.((...)).)))).)).))..))))))..))))). ( -38.90) >DroEre_CAF1 7713 92 - 1 GCCAGGAUGCCACUGCGUGGUGGAAUUUUGGGCAGUGGCGGCUUGCCAGUCACCCUUUGCAGAGUUGAGGUCUUGACCAGCUGUUCCUGUUG .(((((((.(((((....))))).)))))))((((..((((((.....((((.((((.((...)).))))...)))).))))))..)))).. ( -33.60) >DroYak_CAF1 6813 92 - 1 GCCAGGAUGCCACUACGUGGUGGCAUCUUGGGCAGUGGCGGCUUGCCAGUCACCCUUUGCAGGGUUGAGGUCUUGACCAGCUGUUCCUGAGG .(((((((((((((....))))))))))))).(((..((((((......(((((((....)))).)))(((....)))))))))..)))... ( -46.00) >DroAna_CAF1 22155 92 - 1 GCCAGGCUGCCACUUCGGGGAGGCUGCUUGGGCAACGGCGGCUUACCAGUCACCUUCUGCAGUGUGGAGGUUUUCGCCAGCUGUUCUGGCUG .(((.(((((..(....)((((((((((..(....))))))))).))...........))))).)))........(((((.....))))).. ( -36.40) >consensus GCCAGGAUGCCACUUCGUGGUGGCAUCUUGGGCAGUGGCGGCUUGCCAGUCACCCUUUGCAGAGUUGAGGUCUUGCCCAGCUGUUCCUGUUG .(((((((((((((....)))))))))))))((((..((((((.(.(((....((((.((...)).))))..))).).))))))..)))).. (-33.38 = -33.22 + -0.16)



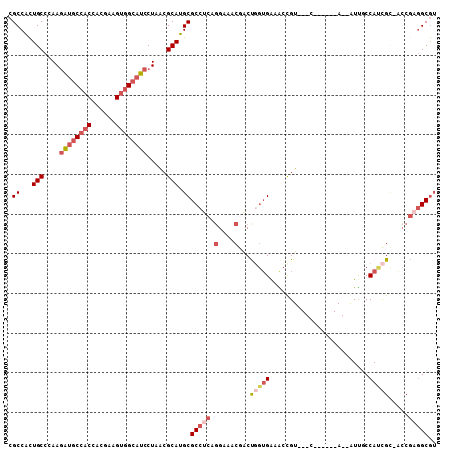
| Location | 22,134,011 – 22,134,107 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -20.59 |
| Energy contribution | -22.26 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22134011 96 + 22407834 CGCCACUGCCCAAGAUGCCACCACGCAGUGGCAUCCUAACGCAUGCGCCUCAGGAAACGACCGGUGAAACUGU---C------A--AUUGCCAUCGC-ACCGAGGCGU ......(((....((((((((......)))))))).....))).(((((((.(....)....((((....((.---(------.--...).))...)-)))))))))) ( -35.10) >DroSec_CAF1 6199 95 + 1 CGCCACUGCCCAAGAUGCCACAACGAAGU-GCAUCCUAACGCAUGCGCCUCAGGAAACGACUGGUGAAACUGU---C------A--GUUGCCACCGC-ACCGAGGCGU ......(((....(((((.((......))-))))).....))).(((((((.(....)....((((.(((...---.------.--)))..))))..-...))))))) ( -30.40) >DroSim_CAF1 6582 96 + 1 CGCCACUGCCCAAGAUGCCACCACGAAGUGGCAUCCUAACGCAUGCGCCUCAGGAAACGACUGGUGAAACUGU---C------A--GUUGCCACCGC-ACCGAGGCGU ......(((....((((((((......)))))))).....))).(((((((.(....)....((((.(((...---.------.--)))..))))..-...))))))) ( -37.00) >DroEre_CAF1 7765 96 + 1 CGCCACUGCCCAAAAUUCCACCACGCAGUGGCAUCCUGGCGCAUGCGCCUCAGGAAACAACCAGUGAAGCCGU---C------A--AUCUCCAUCGU-GCCGUGGCGU .((((((((...............)))))))).((((((((....))))..))))...............(((---(------(--..(........-)...))))). ( -29.46) >DroYak_CAF1 6865 96 + 1 CGCCACUGCCCAAGAUGCCACCACGUAGUGGCAUCCUGGCGCAUGCGCCUCCGGAAACGACUGGUGAAGCCGU---C------A--AUUUCCAUCGC-GCCAUGGCGU (((((.((((((.((((((((......)))))))).))).))).((((....(((((.(((.((.....))))---)------.--.)))))...))-))..))))). ( -44.00) >DroAna_CAF1 22207 108 + 1 CGCCGUUGCCCAAGCAGCCUCCCCGAAGUGGCAGCCUGGCGCAUGCGCCUCACGGAACAGCUCAGGGUGCCGUUGCCGUACCCAGUGUUGCCAGUGAAAGCGAAGCAA ((((((((((((.((.(((.(......).))).)).))).))).)))..((((((..((((.(.((((((.......)))))).).)))))).))))..)))...... ( -37.80) >consensus CGCCACUGCCCAAGAUGCCACCACGAAGUGGCAUCCUAACGCAUGCGCCUCAGGAAACGACUGGUGAAACCGU___C______A__AUUGCCAUCGC_ACCGAGGCGU .((...(((....((((((((......)))))))).....))).))(((((.(....)...(((((.........................))))).....))))).. (-20.59 = -22.26 + 1.67)


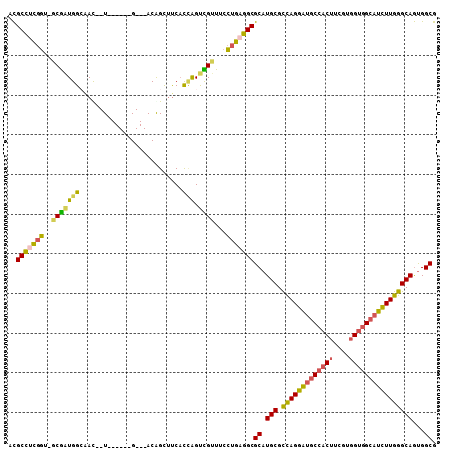
| Location | 22,134,011 – 22,134,107 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -45.62 |
| Consensus MFE | -37.36 |
| Energy contribution | -36.62 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22134011 96 - 22407834 ACGCCUCGGU-GCGAUGGCAAU--U------G---ACAGUUUCACCGGUCGUUUCCUGAGGCGCAUGCGUUAGGAUGCCACUGCGUGGUGGCAUCUUGGGCAGUGGCG ..(((((((.-(.((((((...--(------(---(.....)))...)))))).))))))))((.(((.(((((((((((((....))))))))))))))))...)). ( -46.70) >DroSec_CAF1 6199 95 - 1 ACGCCUCGGU-GCGGUGGCAAC--U------G---ACAGUUUCACCAGUCGUUUCCUGAGGCGCAUGCGUUAGGAUGC-ACUUCGUUGUGGCAUCUUGGGCAGUGGCG ..(((((((.-(((((((.(((--.------.---...)))...))).))))...)))))))((.(((.(((((((((-((......)).))))))))))))...)). ( -37.50) >DroSim_CAF1 6582 96 - 1 ACGCCUCGGU-GCGGUGGCAAC--U------G---ACAGUUUCACCAGUCGUUUCCUGAGGCGCAUGCGUUAGGAUGCCACUUCGUGGUGGCAUCUUGGGCAGUGGCG ..(((((((.-(((((((.(((--.------.---...)))...))).))))...)))))))((.(((.(((((((((((((....))))))))))))))))...)). ( -45.30) >DroEre_CAF1 7765 96 - 1 ACGCCACGGC-ACGAUGGAGAU--U------G---ACGGCUUCACUGGUUGUUUCCUGAGGCGCAUGCGCCAGGAUGCCACUGCGUGGUGGAAUUUUGGGCAGUGGCG .((((((.((-.(...((((((--.------.---.(((.....)))...))))))...)))...(((.(((((((.(((((....))))).)))))))))))))))) ( -40.90) >DroYak_CAF1 6865 96 - 1 ACGCCAUGGC-GCGAUGGAAAU--U------G---ACGGCUUCACCAGUCGUUUCCGGAGGCGCAUGCGCCAGGAUGCCACUACGUGGUGGCAUCUUGGGCAGUGGCG .((((((.((-((..(((((((--.------(---((((.....)).))))))))))...)))).(((.(((((((((((((....)))))))))))))))))))))) ( -55.20) >DroAna_CAF1 22207 108 - 1 UUGCUUCGCUUUCACUGGCAACACUGGGUACGGCAACGGCACCCUGAGCUGUUCCGUGAGGCGCAUGCGCCAGGCUGCCACUUCGGGGAGGCUGCUUGGGCAACGGCG ..(((((((.((((.((....)).))))...((.((((((.......)))))))))))))))((.(((.((((((.(((.(....)...))).)))))))))...)). ( -48.10) >consensus ACGCCUCGGU_GCGAUGGCAAC__U______G___ACAGCUUCACCAGUCGUUUCCUGAGGCGCAUGCGCCAGGAUGCCACUUCGUGGUGGCAUCUUGGGCAGUGGCG ..(((((((..(((((((..........................))).))))...)))))))((.(((.(((((((((((((....))))))))))))))))...)). (-37.36 = -36.62 + -0.74)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:34 2006