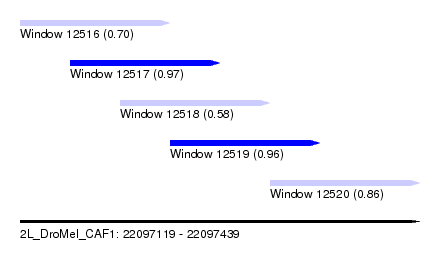
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 22,097,119 – 22,097,439 |
| Length | 320 |
| Max. P | 0.968571 |

| Location | 22,097,119 – 22,097,239 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -36.81 |
| Energy contribution | -36.62 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22097119 120 + 22407834 CAUGCACUUCUAUCUCUUUAAGAGGGAGUUCCCUGAGUGUGGCAGAGAGGUGGAUUUCUCAAUCGAGCGCAAAUGGAGGCAUUGUGGCUUUAAUGCUUCUCUGCCCACACGACUGAUGAU ..........((((........((((....))))..(((((((((((((((.((((....))))((((.((((((....)))).))))))....))))))))).))))))....)))).. ( -38.70) >DroSec_CAF1 4622 120 + 1 CAUGCACUUCUAUCUCUUUAAGAGGGAGUUCCCUGAGUGUGGCAGGGAGGUGGAUUUCUCAAUCGAGCGCAAAUGGAGGCAUUGUGGCUUUAAUGCUUCUCUGCCCACACGAUUGAUGAU ........(((((((((((...((((....))))..(.....))))))))))))..(((((((((.(.(((...((((((((((......)))))))))).)))...).))))))).)). ( -38.60) >DroSim_CAF1 4792 120 + 1 CAUGCACUUCUAUCUCUUUAAGAGGGAGUUCCCUGAGUGUGGCAGGGAGGUGGAUUUCUCAAUCGAGCGCAAAUGGAGGCAUUGUGGCUUUAAUGCUUCUCUGCCCACACGAUUGAUGAU ........(((((((((((...((((....))))..(.....))))))))))))..(((((((((.(.(((...((((((((((......)))))))))).)))...).))))))).)). ( -38.60) >DroYak_CAF1 8456 120 + 1 CAUGCACUUCUACCUCUUUAAGAGGGAGUUCCCAGAGUGUGGCAGGGAGGUGAAUUUCUCAAUCGAGCGAAAGUGGAAGCACUGUGGCUUUAAUGCUUCUCUGCCCACAAGGUUGAUGAU ......(.((.((((........(((....)))...(((.(((((((((((((.....))....((((.(.((((....)))).).))))....)))))))))))))).)))).)).).. ( -42.00) >consensus CAUGCACUUCUAUCUCUUUAAGAGGGAGUUCCCUGAGUGUGGCAGGGAGGUGGAUUUCUCAAUCGAGCGCAAAUGGAGGCAUUGUGGCUUUAAUGCUUCUCUGCCCACACGAUUGAUGAU ......(.((.(((........((((....))))..(((.(((((((((((.((((....))))((((...((((....))))...))))....))))))))))))))..))).)).).. (-36.81 = -36.62 + -0.19)

| Location | 22,097,159 – 22,097,279 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -45.62 |
| Consensus MFE | -43.35 |
| Energy contribution | -43.10 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22097159 120 + 22407834 GGCAGAGAGGUGGAUUUCUCAAUCGAGCGCAAAUGGAGGCAUUGUGGCUUUAAUGCUUCUCUGCCCACACGACUGAUGAUACACGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGG (((((((((((.((((....))))((((.((((((....)))).))))))....)))))))))))..................((((((((.((((((......)))))).)))))))). ( -45.40) >DroSec_CAF1 4662 120 + 1 GGCAGGGAGGUGGAUUUCUCAAUCGAGCGCAAAUGGAGGCAUUGUGGCUUUAAUGCUUCUCUGCCCACACGAUUGAUGAUACACGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGG (((((((((((.((((....))))((((.((((((....)))).))))))....)))))))))))..................((((((((.((((((......)))))).)))))))). ( -44.50) >DroSim_CAF1 4832 120 + 1 GGCAGGGAGGUGGAUUUCUCAAUCGAGCGCAAAUGGAGGCAUUGUGGCUUUAAUGCUUCUCUGCCCACACGAUUGAUGAUACACGGCUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGG (((((((((((.((((....))))((((.((((((....)))).))))))....)))))))))))....(((((((.(((((.(((((((.....))))))).)))..))..))))))). ( -47.50) >DroYak_CAF1 8496 120 + 1 GGCAGGGAGGUGAAUUUCUCAAUCGAGCGAAAGUGGAAGCACUGUGGCUUUAAUGCUUCUCUGCCCACAAGGUUGAUGAUACAUGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGG (((((((((((((.....))....((((.(.((((....)))).).))))....)))))))))))..................((((((((.((((((......)))))).)))))))). ( -45.10) >consensus GGCAGGGAGGUGGAUUUCUCAAUCGAGCGCAAAUGGAGGCAUUGUGGCUUUAAUGCUUCUCUGCCCACACGAUUGAUGAUACACGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGG (((((((((((.((((....))))((((...((((....))))...))))....)))))))))))..................((((((((.((((((......)))))).)))))))). (-43.35 = -43.10 + -0.25)

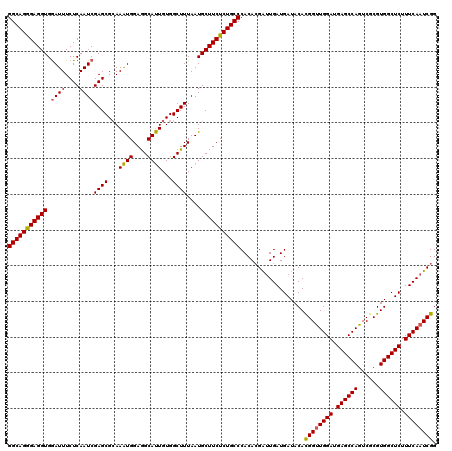
| Location | 22,097,199 – 22,097,319 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -33.86 |
| Energy contribution | -34.17 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22097199 120 + 22407834 AUUGUGGCUUUAAUGCUUCUCUGCCCACACGACUGAUGAUACACGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGGGAUGUGAAGAAUGCAUACCUAAAAAAAGGGGAGUACAAUG ((((((.((((.((((((((..((((.((....))........((((((((.((((((......)))))).))))))))))..))..)))).)))).(((......))))))))))))). ( -36.70) >DroSec_CAF1 4702 120 + 1 AUUGUGGCUUUAAUGCUUCUCUGCCCACACGAUUGAUGAUACACGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGGGAUGUGAAGAAUGCAUACCUAAAAAAAGGGGAGUACAAUG ((((((.((((.((((((((..((...(.(((((((.((..(((((((((.....)))).).))))..))..))))))))...))..)))).)))).(((......))))))))))))). ( -37.00) >DroSim_CAF1 4872 120 + 1 AUUGUGGCUUUAAUGCUUCUCUGCCCACACGAUUGAUGAUACACGGCUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGGGAUGUGAAGAAUGCAUACCUAAAAAAAGGGGAGUACAAUG ((((((.((((.((((((((..((...(.(((((((.(((((.(((((((.....))))))).)))..))..))))))))...))..)))).)))).(((......))))))))))))). ( -40.60) >DroYak_CAF1 8536 120 + 1 ACUGUGGCUUUAAUGCUUCUCUGCCCACAAGGUUGAUGAUACAUGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGGGAUGUGAAAAAUGCAUACCUGAAAAAAGGGGAGUACAAUG ..((((.((((.((((.((((.(((.....))).)).))((((((((((((.((((((......)))))).)))))))...)))))......)))).(((......)))))))))))... ( -33.70) >consensus AUUGUGGCUUUAAUGCUUCUCUGCCCACACGAUUGAUGAUACACGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGGGAUGUGAAGAAUGCAUACCUAAAAAAAGGGGAGUACAAUG ((((((((......))....((.(((.................((((((((.((((((......)))))).))))))))((((((.......)))).))........))).)))))))). (-33.86 = -34.17 + 0.31)

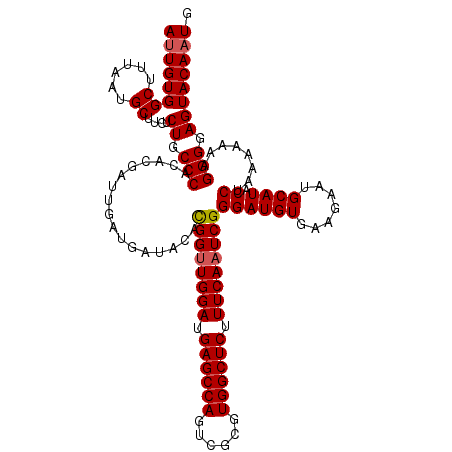
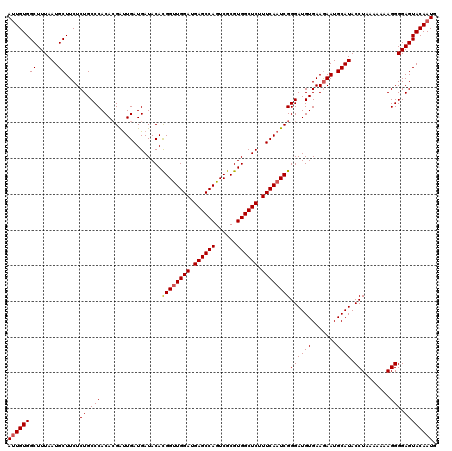
| Location | 22,097,239 – 22,097,359 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -39.15 |
| Consensus MFE | -33.95 |
| Energy contribution | -33.45 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22097239 120 + 22407834 ACACGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGGGAUGUGAAGAAUGCAUACCUAAAAAAAGGGGAGUACAAUGUGAUCGUUGUCGAUUGGAGCGCAAGCUCAGCCAACAUAAA .....(((((..((((((......))))))...((((((((((((.......)))).)))............(.((((((....))))))))))))((((....))))..)))))..... ( -39.70) >DroSec_CAF1 4742 120 + 1 ACACGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGGGAUGUGAAGAAUGCAUACCUAAAAAAAGGGGAGUACAAUGUGAUCGUAGUCGAUUGGAGCGCGAGCUCAGCCAACUUAAA ...((((((((.((((((......)))))).))))))))((((((.......)))).))......(((..(.((.......(((((....))))).((((....)))).)))..)))... ( -38.70) >DroSim_CAF1 4912 120 + 1 ACACGGCUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGGGAUGUGAAGAAUGCAUACCUAAAAAAAGGGGAGUACAAUGUGAUCGUAGUCGAUUGGAGCGCGAGCUCAGCCAACUUAAA ((.(((((((.....))))))).))((((....((((((((((((.......)))).)))..........((.(((.........))).)))))))((((....))))))))........ ( -40.30) >DroYak_CAF1 8576 120 + 1 ACAUGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGGGAUGUGAAAAAUGCAUACCUGAAAAAAGGGGAGUACAAUGUGAUCGUUGCCGAUUGGAGUUCAAGUUCAGCUAAUAUAAA ...(((((((((((((((......))))))((((((((((...........(((...(((......)))...)))(((((....))))))))))))))).....)))))))))....... ( -37.90) >consensus ACACGGUUGGAUGAGCCAGUCGCGUGGCUCUUUCAAUCGGGAUGUGAAGAAUGCAUACCUAAAAAAAGGGGAGUACAAUGUGAUCGUAGUCGAUUGGAGCGCAAGCUCAGCCAACAUAAA ...((((((((.((((((......)))))).))))))))((((((.......)))).))........((............(((((....))))).((((....))))..))........ (-33.95 = -33.45 + -0.50)

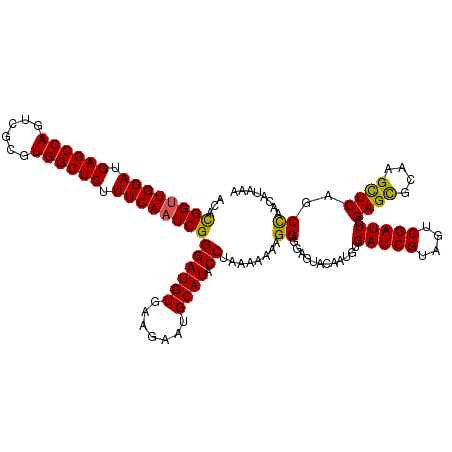
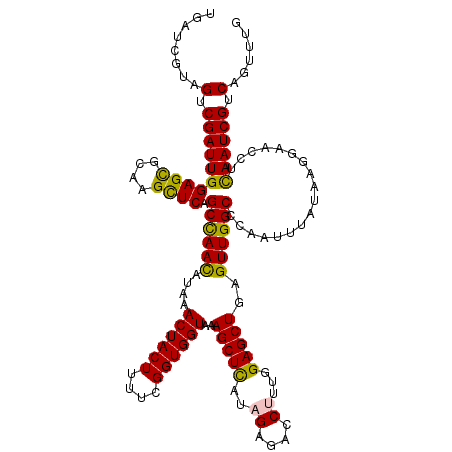
| Location | 22,097,319 – 22,097,439 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -32.89 |
| Energy contribution | -32.20 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22097319 120 + 22407834 UGAUCGUUGUCGAUUGGAGCGCAAGCUCAGCCAACAUAAACUACUUUUCGGUGGUAAAGCUUAUAGAGACCUUUGGAGCUGAGUUGGCCCAAUUUAUAAGGAACCUCAAUCGUCAGUUUG ......(((.((((((((((....)))).((((((....((((((....))))))..(((((..((....))...)))))..))))))..................)))))).))).... ( -38.90) >DroSec_CAF1 4822 120 + 1 UGAUCGUAGUCGAUUGGAGCGCGAGCUCAGCCAACUUAAACUACUUUUCGGUGGUGAAGCUCAUAGAGACCUUUGGAGCUGAGUUGGCCCAAUUUAUAAGGAACCUCAAUCGUCAGUUUG ........(.((((((((((....)))).((((((((..((((((....))))))..(((((.((((....)))))))))))))))))..................)))))).)...... ( -41.00) >DroSim_CAF1 4992 120 + 1 UGAUCGUAGUCGAUUGGAGCGCGAGCUCAGCCAACUUAAACUACUUUUCGGUGGUGAAGCUCAUUGAGACCUUUGGAGCUGAGUUGGCCCAAUUUAUAAGGAACCUCAAUCGUCAGUUUG ........(.((((((((((....)))).((((((((..((((((....))))))..(((((...((....))..)))))))))))))..................)))))).)...... ( -39.90) >DroYak_CAF1 8656 120 + 1 UGAUCGUUGCCGAUUGGAGUUCAAGUUCAGCUAAUAUAAACUACUUUUCGGUGGUAAAGCUUAUAGAGACCCUUGGAGCUCAGUUGGCCCAAUUUAUAAGGGACCUUAAUCGUCAGUUUG ......(((.((((((..((((.................((((((....))))))....((((((((.....((((.(((.....)))))))))))))))))))..)))))).))).... ( -27.40) >consensus UGAUCGUAGUCGAUUGGAGCGCAAGCUCAGCCAACAUAAACUACUUUUCGGUGGUAAAGCUCAUAGAGACCUUUGGAGCUGAGUUGGCCCAAUUUAUAAGGAACCUCAAUCGUCAGUUUG ........(.((((((((((....)))).((((((....((((((....))))))..(((((..((....))...)))))..))))))..................)))))).)...... (-32.89 = -32.20 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:25 2006