
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 22,092,679 – 22,092,778 |
| Length | 99 |
| Max. P | 0.620266 |

| Location | 22,092,679 – 22,092,778 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 64.57 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -15.34 |
| Energy contribution | -14.95 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 22092679 99 - 22407834 GUCCUGGCUAUCGCAGUCCCUUGCGAUACACAAAAAUGCUUCGUGAGAUGAACUCUCGCAACGGGCUUUAGUAUGUAUAUAU---ACAA--------------UAUAUUCCU---CGAA ((((((..((((((((....))))))))..))..........((((((.....))))))...))))....(((((....)))---))..--------------.........---.... ( -24.80) >DroVir_CAF1 4805 119 - 1 GUCCCGGCUACCGCAGCCCACUGCGAUACACAAAAGUCUUAAGAGAAAUGAACUCGCGCAACGGACUCUAGUAGGUGCAUCCGAUAUUUUAGUCCAAUCUUAAUAUGGAAUACCCUCUU ((((..((...(((((....))))).................(((.......)))..))...))))...((..((((..(((.(((((..((......)).)))))))).))))..)). ( -25.60) >DroPse_CAF1 27933 112 - 1 GACCCGGCUAUCGCAGUCCCCUGCGAUACAAUAAGAUGCUCAAAGAGAUGAACUCUCGCAAUGGUCUUUAGUAAGUCUAA-G---AUCUUGG---GCACAUACUAGCUCCCACGUUUAU ((((..((((((((((....))))))))................((((.....))))))...)))).((((((.((((((-.---...))))---))...))))))............. ( -29.90) >DroGri_CAF1 18334 108 - 1 GUCCCGGCUAUCGCAGCCCCCUGCGUUACACCAAGGUCAUGCGAGAGAUGAACUCUCGCAACGGACUCUAGUAAGAAAAACUGUUAUUUG-------UUUGA----AGAGCACCCCCUU ((((.((....(((((....))))).....)).......((((((((.....))))))))..))))....((.....((((........)-------)))..----...))........ ( -28.80) >DroAna_CAF1 163 105 - 1 GCCCCGGAUAUCGUAGUCCUCUACGUUAUCAGAAGAUCAUGCGCGAAAUGAACUCUCGCAACGGGCUCUAGUAAGUCCACAU---AUCAU------CACAU--CUUAGUCCU---UUAU ......((((.(((((....))))).)))).((.(((.(((.((((.........))))...(((((......))))).)))---))).)------)....--.........---.... ( -23.60) >DroPer_CAF1 28173 112 - 1 GACCCGGCUAUCGCAGUCCCCUGCGAUACAAUAAGAUGCUCAAAGAGAUGAACUCUCGCAAUGGUCUUUAGUAAGUCUAA-G---AUCUUGG---GUAUUUACUAGCUACCACGUUUAU .....(((((((((((....)))))).......((((((((((.((((.....)))).....((((((.((.....))))-)---)))))))---))))))..)))))........... ( -31.10) >consensus GUCCCGGCUAUCGCAGUCCCCUGCGAUACAAAAAGAUCCUCAGAGAGAUGAACUCUCGCAACGGACUCUAGUAAGUCUAA_U___AUCUU_______ACUUA_UAUAGACCAC__UUAU ((((..((...(((((....)))))...................((((.....))))))...))))..................................................... (-15.34 = -14.95 + -0.39)

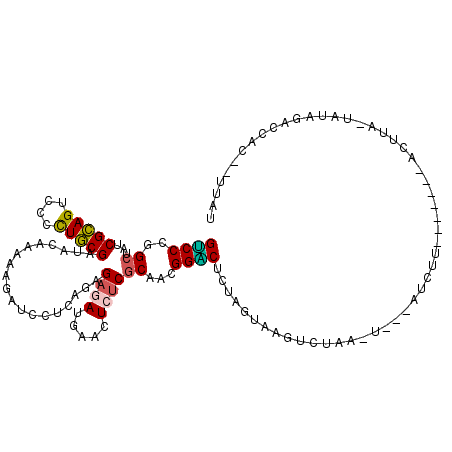

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:21 2006