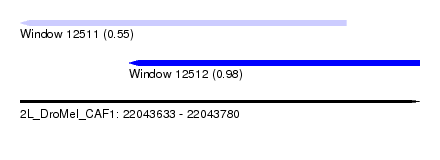
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 22,043,633 – 22,043,780 |
| Length | 147 |
| Max. P | 0.981386 |

| Location | 22,043,633 – 22,043,753 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -24.41 |
| Energy contribution | -24.52 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
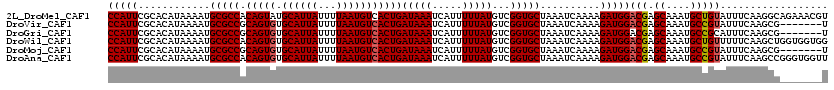
>2L_DroMel_CAF1 22043633 120 - 22407834 CCAUUCGCACAUAAAAUGCGCCACAGUAUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAAAAGAUGGACGAGCAAAUGCUGUAUUUCAAGGCAGAAACGU ...(((((.........(((((.((((..((((((...)))))).))))(((((.....)))))...)))))..........((...((.(((....)))))...))...)).))).... ( -23.40) >DroVir_CAF1 73511 113 - 1 CCAUUCGCACAUAAAAUGCGCCGCAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAAAAGAUGGACGAGCAAAUGCCGUAUUUCAAGCG-------U .....(((.........(((((((((((.((((((...)))))))))))(((((.....)))))..))))))..........((...(((.((....)))))...))..)))-------. ( -29.80) >DroGri_CAF1 82472 113 - 1 CCAUUCGCACAUAAAAUGCGCCGCAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAAAAGAUGGACGAGCAAAUGCCGCAUUUCAAGCG-------U .....(((.........(((((((((((.((((((...)))))))))))(((((.....)))))..))))))..........((..(.((.((....)))))...))..)))-------. ( -27.60) >DroWil_CAF1 54380 120 - 1 CCAUUCGCACAUAAAAUGCGCCACAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAAAAGAUGGACGAGCAAAUGCUGUUUUUCAAGCUGGUGGUGG ((((..((.........(((((.(((((.((((((...)))))))))))(((((.....)))))...)))))..........((..(((.(((....))))))..))..))..))))... ( -33.90) >DroMoj_CAF1 88448 113 - 1 CCAUUCGCACAUAAAAUGCGCCGCAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAAAAGAUGGACGAGCAAAUGCCGUAUUUCAAGCG-------U .....(((.........(((((((((((.((((((...)))))))))))(((((.....)))))..))))))..........((...(((.((....)))))...))..)))-------. ( -29.80) >DroAna_CAF1 78066 120 - 1 CCAUUCGCACAUAAAAUGCGCCACAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAAAAGAUGGACGAGCAAAUGCCGUAUUUCAAGCCGGGUGGUU ((((((((.........(((((.(((((.((((((...)))))))))))(((((.....)))))...)))))..........((...(((.((....)))))...))..).))))))).. ( -34.10) >consensus CCAUUCGCACAUAAAAUGCGCCACAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAAAAGAUGGACGAGCAAAUGCCGUAUUUCAAGCG_______U (((((............(((((.(((((.((((((...)))))))))))(((((.....)))))...)))))..........)))))(((.((....))))).................. (-24.41 = -24.52 + 0.11)


| Location | 22,043,673 – 22,043,780 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -24.56 |
| Energy contribution | -24.58 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
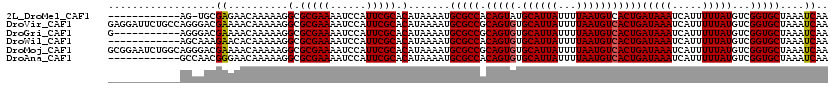
>2L_DroMel_CAF1 22043673 107 - 22407834 ------------AG-UGCGAGAACAAAAAGGCGCGAAAAUCCAUUCGCACAUAAAAUGCGCCACAGUAUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAA ------------.(-((((((......................))))))).......(((((.((((..((((((...)))))).))))(((((.....)))))...)))))........ ( -22.95) >DroVir_CAF1 73544 120 - 1 GAGGAUUCUGCCAGGGACGAAAACAAAAAGGCGCGAAAAUCCAUUCGCACAUAAAAUGCGCCGCAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAA ...((((..(((.................)))(((((......))))).........(((((((((((.((((((...)))))))))))(((((.....)))))..))))))..)))).. ( -29.43) >DroGri_CAF1 82505 109 - 1 G-----------AGGGACGAAAACAAAAAGGCGCGAAAAUCCAUUCGCACAUAAAAUGCGCCGCAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAA .-----------......((..........(.(((((......))))).).......(((((((((((.((((((...)))))))))))(((((.....)))))..))))))....)).. ( -26.10) >DroWil_CAF1 54420 108 - 1 ------------AGCAAAGAACACAAAAAGGCGCGAAAAUCCAUUCGCACAUAAAAUGCGCCACAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAA ------------((((..((.((.(((((((((((((......))))).(((...))).))).(((((.((((((...)))))))))))........))))).))))..))))....... ( -28.20) >DroMoj_CAF1 88481 120 - 1 GCGGAAUCUGGCAGGGACGAAAACAAAAAGGCGCGAAAAUCCAUUCGCACAUAAAAUGCGCCGCAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAA ........(((((..((((..........((((((((......))))).(((...))).))).(((((.((((((...)))))))))))..............))))..)))))...... ( -29.00) >DroAna_CAF1 78106 108 - 1 ------------GCCAACGGGAACAAAAAGGCGCGAAAAUCCAUUCGCACAUAAAAUGCGCCACAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAA ------------(((...(....).....)))(((((......))))).........(((((.(((((.((((((...)))))))))))(((((.....)))))...)))))........ ( -28.40) >consensus ____________AGGGACGAAAACAAAAAGGCGCGAAAAUCCAUUCGCACAUAAAAUGCGCCACAGUGUGCAUUAUUUUAAUGUCACUGAUAAAUCAUUUUUAUGUCGGUGCUAAAUCAA ..................((..........(.(((((......))))).).......(((((.(((((.((((((...)))))))))))(((((.....)))))...)))))....)).. (-24.56 = -24.58 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:18 2006