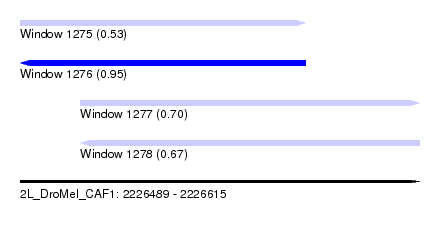
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,226,489 – 2,226,615 |
| Length | 126 |
| Max. P | 0.948026 |

| Location | 2,226,489 – 2,226,579 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -18.28 |
| Energy contribution | -19.40 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2226489 90 + 22407834 -----CGUUAUCCAACACUUAAGGUCUGGAAAAUUGCCAGUACUGAAAAGUGCGGAAAUCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUA -----.(((.(((..(((((..(((((((.......)))).)))...))))).))).....((((((((....)))).))))...)))....... ( -22.90) >DroSec_CAF1 8571 90 + 1 -----CGUUAUCCAACACUUAAGGUCUGGAAAAUUGCCAGUGCUGGAAAAGGCGGAAAGCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUA -----.(((.(((............((((.......)))).(((......)))))).))).((((..(((((....)))))....))))...... ( -19.90) >DroSim_CAF1 7586 90 + 1 -----CGUUAUCCAACACUUAAGGUCUGGAAAAUUGCCAGUGCUGGAAAAGGCGGAAAGCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUA -----.(((.(((............((((.......)))).(((......)))))).))).((((..(((((....)))))....))))...... ( -19.90) >DroEre_CAF1 7747 95 + 1 CGCAACGUUGUCCAACACUAAGGGUCUGGAAAAUUGCCAGUGCUGGAAAGUGCGGAAAGCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUA ((((.(....((((.((((...(((..........))))))).))))..))))).......((((..(((((....)))))....))))...... ( -22.00) >DroYak_CAF1 7733 95 + 1 CGCAACGUUUUCCAACACUAAGAGCCUGGAAAAUAGCCAGUGGUGGAAAGUGCGGAAAGCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUA ......(((((((..((((....((((((.......)))..)))....)))).))))))).((((..(((((....)))))....))))...... ( -24.60) >consensus _____CGUUAUCCAACACUUAAGGUCUGGAAAAUUGCCAGUGCUGGAAAGUGCGGAAAGCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUA ......(((.(((..(((((..(((((((.......)))).)))...))))).))).))).((((..(((((....)))))....))))...... (-18.28 = -19.40 + 1.12)


| Location | 2,226,489 – 2,226,579 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -20.72 |
| Energy contribution | -20.44 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2226489 90 - 22407834 UAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGAUUUCCGCACUUUUCAGUACUGGCAAUUUUCCAGACCUUAAGUGUUGGAUAACG----- ...(..((((...(((((......))))).))))..)..(((((((((....((.((((.......))))))...))))).)))).....----- ( -25.60) >DroSec_CAF1 8571 90 - 1 UAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGCUUUCCGCCUUUUCCAGCACUGGCAAUUUUCCAGACCUUAAGUGUUGGAUAACG----- .....(((((...(((((......))))).))))).............((((((((((((.........).))...))))))))).....----- ( -22.90) >DroSim_CAF1 7586 90 - 1 UAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGCUUUCCGCCUUUUCCAGCACUGGCAAUUUUCCAGACCUUAAGUGUUGGAUAACG----- .....(((((...(((((......))))).))))).............((((((((((((.........).))...))))))))).....----- ( -22.90) >DroEre_CAF1 7747 95 - 1 UAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGCUUUCCGCACUUUCCAGCACUGGCAAUUUUCCAGACCCUUAGUGUUGGACAACGUUGCG .....(((((...(((((......))))).)))))......((((((((((((((((((...............))))))))))).)).).)))) ( -25.96) >DroYak_CAF1 7733 95 - 1 UAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGCUUUCCGCACUUUCCACCACUGGCUAUUUUCCAGGCUCUUAGUGUUGGAAAACGUUGCG .....(((((...(((((......))))).)))))......((((.((((((.((((((((........))))...)))).))))))....)))) ( -27.70) >consensus UAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGCUUUCCGCACUUUCCAGCACUGGCAAUUUUCCAGACCUUAAGUGUUGGAUAACG_____ .....(((((...(((((......))))).))))).............((((((((((((.........).))...))))))))).......... (-20.72 = -20.44 + -0.28)

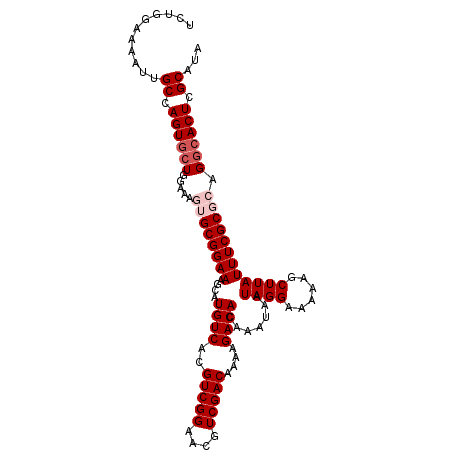
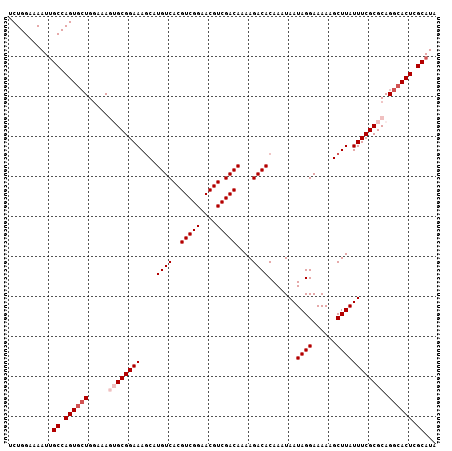
| Location | 2,226,508 – 2,226,615 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -23.92 |
| Energy contribution | -25.12 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2226508 107 + 22407834 UCUGGAAAAUUGCCAGUACUGAAAAGUGCGGAAAUCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUAAUAGGAAAAAGCUUAUUUCGCGCAGGCACUCGCAUA .((((.......)))).........((((((((....((((..(((((....)))))....)))).......((((......))))))))))))..((....))... ( -23.40) >DroSec_CAF1 8590 107 + 1 UCUGGAAAAUUGCCAGUGCUGGAAAAGGCGGAAAGCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUAAUAGGAAAAAGCUUAUUUCGCGCAGGCACUCGCAUA ..........(((.((((((.(.....(((((((((.((((..(((((....)))))....))))(........)......)))..)))))).).)))))).))).. ( -26.00) >DroSim_CAF1 7605 107 + 1 UCUGGAAAAUUGCCAGUGCUGGAAAAGGCGGAAAGCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUAAUAGGAAAAAGCUUAUUUCGCGCAGGCACUCGCAUA ..........(((.((((((.(.....(((((((((.((((..(((((....)))))....))))(........)......)))..)))))).).)))))).))).. ( -26.00) >DroEre_CAF1 7771 107 + 1 UCUGGAAAAUUGCCAGUGCUGGAAAGUGCGGAAAGCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUAAUAGGAAAAAGCUUAUUUCGCGCAGGCACUCGCAUA ..........(((.((((((.....(((((((((((.((((..(((((....)))))....))))(........)......)))..)))))))).)))))).))).. ( -30.30) >DroYak_CAF1 7757 107 + 1 CCUGGAAAAUAGCCAGUGGUGGAAAGUGCGGAAAGCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUAAUAGGAAAAAGCUUAUUUCGCGCAGGCACUCGCAUA ..(((.......)))((((((....(((((((((((.((((..(((((....)))))....))))(........)......)))..))))))))...))).)))... ( -25.40) >consensus UCUGGAAAAUUGCCAGUGCUGGAAAGUGCGGAAAGCAUGUCACGUCGGAACGUCGACAAAAGACACAAAUAAUAGGAAAAAGCUUAUUUCGCGCAGGCACUCGCAUA ...........((.((((((.....((((((((....((((..(((((....)))))....)))).......((((......)))))))))))).)))))).))... (-23.92 = -25.12 + 1.20)

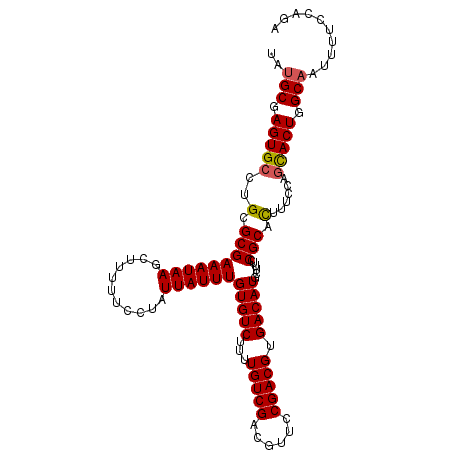
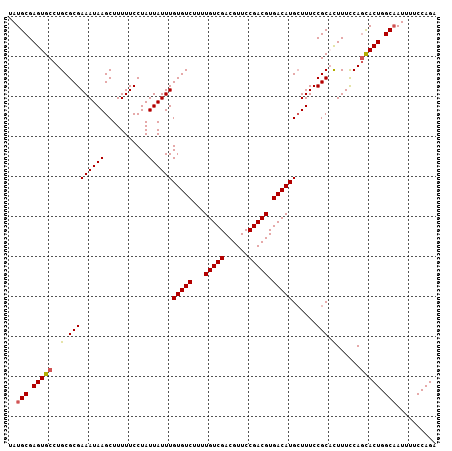
| Location | 2,226,508 – 2,226,615 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -24.72 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2226508 107 - 22407834 UAUGCGAGUGCCUGCGCGAAAUAAGCUUUUUCCUAUUAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGAUUUCCGCACUUUUCAGUACUGGCAAUUUUCCAGA ..(((.(((((.((((.(((((.((.......)).......(((((...(((((......))))).))))).))))))))).......))))).))).......... ( -26.00) >DroSec_CAF1 8590 107 - 1 UAUGCGAGUGCCUGCGCGAAAUAAGCUUUUUCCUAUUAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGCUUUCCGCCUUUUCCAGCACUGGCAAUUUUCCAGA ..(((.(((((.((.(((....((((...............(((((...(((((......))))).)))))))))..)))......))))))).))).......... ( -26.56) >DroSim_CAF1 7605 107 - 1 UAUGCGAGUGCCUGCGCGAAAUAAGCUUUUUCCUAUUAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGCUUUCCGCCUUUUCCAGCACUGGCAAUUUUCCAGA ..(((.(((((.((.(((....((((...............(((((...(((((......))))).)))))))))..)))......))))))).))).......... ( -26.56) >DroEre_CAF1 7771 107 - 1 UAUGCGAGUGCCUGCGCGAAAUAAGCUUUUUCCUAUUAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGCUUUCCGCACUUUCCAGCACUGGCAAUUUUCCAGA ..(((.(((((.((((.(((...(((...............(((((...(((((......))))).))))))))))))))).......))))).))).......... ( -27.66) >DroYak_CAF1 7757 107 - 1 UAUGCGAGUGCCUGCGCGAAAUAAGCUUUUUCCUAUUAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGCUUUCCGCACUUUCCACCACUGGCUAUUUUCCAGG .....((((((..((((.......))...............(((((...(((((......))))).))))))).....)))))).......((((.......)))). ( -24.60) >consensus UAUGCGAGUGCCUGCGCGAAAUAAGCUUUUUCCUAUUAUUUGUGUCUUUUGUCGACGUUCCGACGUGACAUGCUUUCCGCACUUUCCAGCACUGGCAAUUUUCCAGA ..(((.(((((..(.(((((((((...........))))))(((((...(((((......))))).)))))......))).)......))))).))).......... (-24.72 = -24.72 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:27 2006