| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 22,009,346 – 22,009,454 |
| Length | 108 |
| Max. P | 0.650340 |

| Location | 22,009,346 – 22,009,454 |
|---|---|
| Length | 108 |
| Sequences | 6 |
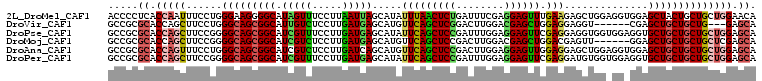
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.21 |
| Mean single sequence MFE | -36.44 |
| Consensus MFE | -27.98 |
| Energy contribution | -27.02 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
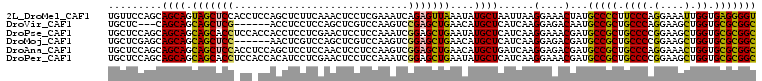
>2L_DroMel_CAF1 22009346 108 + 22407834 UGUUCCAGCAGCAGUAGCUCCACCUCCAGCUCUUCAAACUCCUCGAAAUCAGAGUUAAAUAUGCUAAUUAAGGAAACUAUGCCCCUUCCCAGGAAAUUGGUGAGGGGU ......(((((.((.((((........)))))).).(((((..........))))).....)))).....((....))..((((((..((((....))))..)))))) ( -29.10) >DroVir_CAF1 41170 99 + 1 UGCUC---CAGCAGCAGCUCG------ACCUCCUCCAGCUCGUCCAAGUCCGAGCUGAACAUGCUCAUCAAGGAGACAAUGCCGCUGCCCAGGAAGCUGGUGCGCGGC (((((---(((((.(((((((------..((...............))..)))))))....))))......)))).))..(((((.((((((....)))).))))))) ( -39.36) >DroPse_CAF1 70272 108 + 1 UGCUCCAGCAGCAGCAGCACCUCCACCACCUCCUCGAACUCCUCCAAAUCGGAGCUGAAUAUGCUCAUCAAGGAAACGAUGCCGCUGCCCCGGAAGCUGGUGCGCGGC .((.(((((.(((((.(((.((((..........(((...........)))((((.......)))).....)))...).))).))))).......))))).))..... ( -30.90) >DroMoj_CAF1 55281 102 + 1 UGCUCGAGCAGCAGCAGCUCC------AACUCGUCCAGCUCGUCCAAGUCGGAGCUGAACAUGCUCAUCAAGGAGACGAUGCCGCUGCCCCGGAAGCUGGUGCGCGGC .....(((((....(((((((------.(((.(..(.....)..).))).)))))))....))))).....(....)...(((((.((.((((...)))).))))))) ( -42.40) >DroAna_CAF1 51003 108 + 1 UGCUCCAGCAGCAGCAGCUCCACCUCCAGCUCCUCCAACUCCUCCAAGUCGGAGCUGAACAUGCUGAUCAAGGAGACGAUGCCGCUGCCCAGGAAACUGGUGCGCGGC ..((((..(((((.(((((((..((.....................))..)))))))....))))).....)))).....(((((.((((((....)))).))))))) ( -45.60) >DroPer_CAF1 71079 108 + 1 UGCUCCAGCAGCAGCAGCACCUCCACCACAUCCUCGAACUCCUCCAAAUCGGAGCUGAAUAUGCUCAUCAAGGAAACGAUGCCGCUGCCCCGGAAGCUGGUGCGCGGC .((.(((((..(.(((((..........(((..(((..((((........)))).)))..)))..((((..(....)))))..)))))....)..))))).))..... ( -31.30) >consensus UGCUCCAGCAGCAGCAGCUCC_CC_CCACCUCCUCCAACUCCUCCAAAUCGGAGCUGAACAUGCUCAUCAAGGAAACGAUGCCGCUGCCCAGGAAGCUGGUGCGCGGC .........((((.(((((((.............................)))))))....))))......(....)...(((((.((((.(....).)).))))))) (-27.98 = -27.02 + -0.97)
| Location | 22,009,346 – 22,009,454 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.21 |
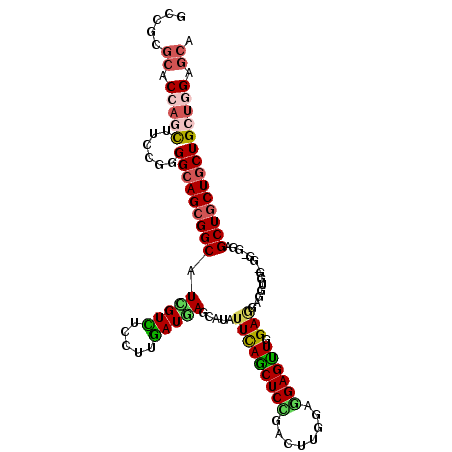
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -31.36 |
| Energy contribution | -31.53 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
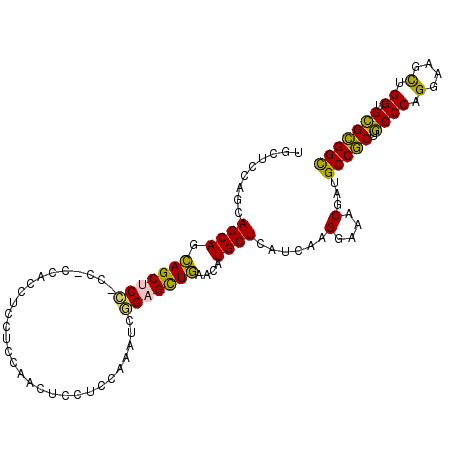
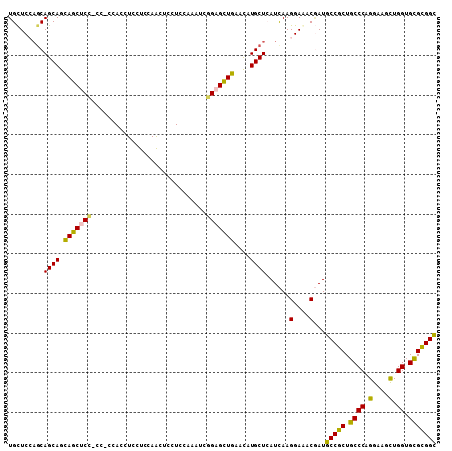
>2L_DroMel_CAF1 22009346 108 - 22407834 ACCCCUCACCAAUUUCCUGGGAAGGGGCAUAGUUUCCUUAAUUAGCAUAUUUAACUCUGAUUUCGAGGAGUUUGAAGAGCUGGAGGUGGAGCUACUGCUGCUGGAACA ..((((..(((......)))..))))(((((((((((((...((((...(((((((((........)))).)))))..)))).))).))))))).))).......... ( -32.50) >DroVir_CAF1 41170 99 - 1 GCCGCGCACCAGCUUCCUGGGCAGCGGCAUUGUCUCCUUGAUGAGCAUGUUCAGCUCGGACUUGGACGAGCUGGAGGAGGU------CGAGCUGCUGCUG---GAGCA ...........(((((...(((((((((.(((((((((((((......)))(((((((........)))))))))))))).------)))))))))))))---)))). ( -45.40) >DroPse_CAF1 70272 108 - 1 GCCGCGCACCAGCUUCCGGGGCAGCGGCAUCGUUUCCUUGAUGAGCAUAUUCAGCUCCGAUUUGGAGGAGUUCGAGGAGGUGGUGGAGGUGCUGCUGCUGCUGGAGCA .....((.(((((..(.(.((((.(..(((((..(((((((((((....))))(((((........))))))))))))..)))))..).)))).).)..))))).)). ( -47.90) >DroMoj_CAF1 55281 102 - 1 GCCGCGCACCAGCUUCCGGGGCAGCGGCAUCGUCUCCUUGAUGAGCAUGUUCAGCUCCGACUUGGACGAGCUGGACGAGUU------GGAGCUGCUGCUGCUCGAGCA (((((......)).....((((((((((.(((((.....)))))........((((((((((((..(.....)..))))))------))))))))))))))))).)). ( -50.20) >DroAna_CAF1 51003 108 - 1 GCCGCGCACCAGUUUCCUGGGCAGCGGCAUCGUCUCCUUGAUCAGCAUGUUCAGCUCCGACUUGGAGGAGUUGGAGGAGCUGGAGGUGGAGCUGCUGCUGCUGGAGCA .....((.(((((......(((((((((..(..((((......(((.(.(((((((((........))))))))).).)))))))..)..)))))))))))))).)). ( -52.60) >DroPer_CAF1 71079 108 - 1 GCCGCGCACCAGCUUCCGGGGCAGCGGCAUCGUUUCCUUGAUGAGCAUAUUCAGCUCCGAUUUGGAGGAGUUCGAGGAUGUGGUGGAGGUGCUGCUGCUGCUGGAGCA .....((.(((((......((((((((((((...(((.......((((.(((((((((........)))))).))).))))...)))))))))))))))))))).)). ( -47.40) >consensus GCCGCGCACCAGCUUCCGGGGCAGCGGCAUCGUCUCCUUGAUGAGCAUAUUCAGCUCCGACUUGGAGGAGUUGGAGGAGGUGG_GG_GGAGCUGCUGCUGCUGGAGCA .....((.(((((......(((((((((.(((((.....))))).....(((((((((........)))))).)))..............)))))))))))))).)). (-31.36 = -31.53 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:08 2006