
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,964,000 – 21,964,115 |
| Length | 115 |
| Max. P | 0.706904 |

| Location | 21,964,000 – 21,964,115 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -19.97 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21964000 115 + 22407834 CGGUUAUUUAUUUUUGCUUCAGUGCGAAGUGUCACCGACUUCGCAGCGCUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAA-ACC-UC--C-UCGGGGCUGCCGUCCGAUUACA .((((((((((.((((......((((((((.......))))))))(((...............)))..))))..))))))))))...-...-..--.-...((((....))))....... ( -28.16) >DroVir_CAF1 4560 100 + 1 CGGCUAUUUAUUUUUGC-GCACUGCCAAGUGUCACCGACUUUGCGGCGAUUCCAUCAAUCAAUCGCGUCGAAUUAUAAAUAACCAAC-GAU-----A-UUGCGACUG-C----------- .((.(((((((.(((((-((.((((.((((.......)))).)))).((((.....))))....))).))))..))))))).))...-...-----.-.........-.----------- ( -22.10) >DroPse_CAF1 34075 114 + 1 CGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAAAACC-UC--CAUCGGGG---CCGACCGAUUGCC .((((((((((.((((......(((.((((.......)))).)))((((((.........))))))..))))..)))))))))).......-..--.(((((..---....))))).... ( -28.10) >DroMoj_CAF1 17115 105 + 1 CGGCUAUUUAUUUUUGC-GCACUGCCAAGUGUCACCGACUUUGCGGCGAUUCCAUCAAUCAAUCGCAUCGAAUUAUAAAUAACCAAC-GACGUGACG-UUCCAACUC-C----------- .((.(((((((..((..-(((((....)))))...(((...(((((.((((.....))))..))))))))))..))))))).))(((-(......))-)).......-.----------- ( -19.80) >DroAna_CAF1 20126 115 + 1 CGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUCCGAAUUAUAAAUAACCAAA-ACC-AC--C-UCGGGGCCGCCAUCCGAUUGCC .((((((((((.((((......(((.((((.......)))).)))((((((.........))))))..))))..))))))))))...-...-..--.-(((((.......)))))..... ( -27.00) >DroPer_CAF1 33926 114 + 1 CGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAAAACC-UC--CAUCGGGG---CCGACCGAUUGCC .((((((((((.((((......(((.((((.......)))).)))((((((.........))))))..))))..)))))))))).......-..--.(((((..---....))))).... ( -28.10) >consensus CGGUUAUUUAUUUUUGCCUCAGUGCCAAGUGUCACCGACUUUGCAGCGAUUCCAUCAAUCAAUCGCUUCGAAUUAUAAAUAACCAAA_ACC_UC__C_UCGGGGCUGCCG_CCGAUUGCC .((((((((((.((((......(((.((((.......)))).)))((((((.........))))))..))))..)))))))))).................................... (-19.97 = -20.25 + 0.28)
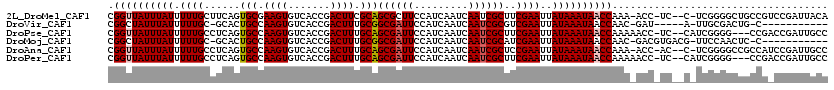
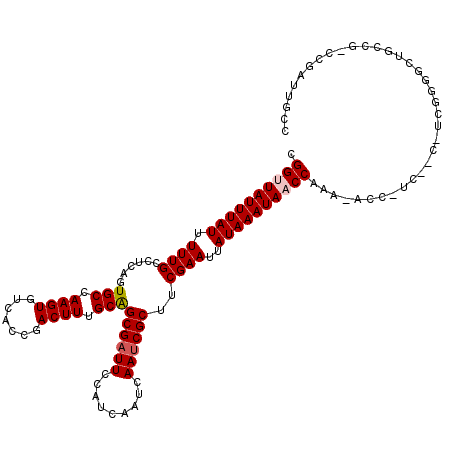

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:59 2006