
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,950,539 – 21,950,629 |
| Length | 90 |
| Max. P | 0.886652 |

| Location | 21,950,539 – 21,950,629 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.18 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -9.73 |
| Energy contribution | -9.35 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
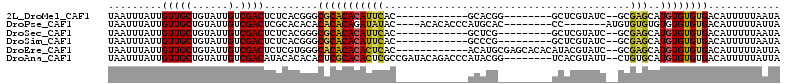
>2L_DroMel_CAF1 21950539 90 + 22407834 UAAUUUAUUGUUGCUGUAUUGUCGACUCUCACGGGCGCACACAUUCAC------------GCACGG--------GCUCGUAUC--GCGAGCAUGUGUGUGACAUUUUUAAUA .....(((((..(.(((..(((((.(((....))))).)))....(((------------(((((.--------(((((....--.))))).))))))))))).)..))))) ( -27.20) >DroPse_CAF1 15095 93 + 1 UAAUUUAUUGUUGCUGUAUUGUCGACUCGCACACACACACAGAUAUAC----ACACACCCAUGCAC--------CC-------AUGUGUGUGUGUGUGUGACAUUUUUAUUA .........(((((......).))))(((((((((((((((.((....----.((......))...--------..-------)).)))))))))))))))........... ( -27.40) >DroSec_CAF1 522 89 + 1 UAAUUUAUUGUUGCUGUAUUGUCGACUCUCACGGGCGCACACAUUCAC------------GCUCG---------GCUCGUAUC--GCGAGCAUGUGUGUGACAUUUUUAAUA ...................((((.((.(.((((((((..........)------------)))))---------(((((....--.))))).)).).))))))......... ( -22.70) >DroSim_CAF1 3759 89 + 1 UAAUUUAUUGUUGCUGUAUUGUCGACUCUCACGGGCGCACACAUUCAC------------GCCCG---------GCUCGUAUC--GCGAGCAUGUGUGUGACAUUUUUAAUA ...................((((.((.(.((((((((..........)------------)))))---------(((((....--.))))).)).).))))))......... ( -25.70) >DroEre_CAF1 2490 98 + 1 UAAUUUAUUGUUGCUGUAUUGUCGACUCUCGUGGGCACACACACUCAC------------ACAUGCGAGCACACAUACGUAUC--GCGAGCAUGUGUGUGACAUUUUUAUUA ........(((..((((..((((.((....)).)))).))).....((------------((((((..((..((....))...--))..)))))))))..)))......... ( -25.80) >DroAna_CAF1 6677 102 + 1 UAAUUUAUUGUUGCUGUAUUGUCGACAUACACACACUCGCACACUCGCCGAUACAGACCCAUACGG--------UCACGUAUU--CUGUGCAUGUGUGUGACAUUUUUAUUA ........((((((......).)))))..(((((((..(((((......(((((.((((.....))--------))..)))))--.)))))..)))))))............ ( -27.80) >consensus UAAUUUAUUGUUGCUGUAUUGUCGACUCUCACGGGCGCACACAUUCAC____________ACACG_________GCUCGUAUC__GCGAGCAUGUGUGUGACAUUUUUAAUA .........(((((......).)))).........(((((((((((.........................................)))..))))))))............ ( -9.73 = -9.35 + -0.39)


| Location | 21,950,539 – 21,950,629 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.18 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -7.82 |
| Energy contribution | -8.88 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
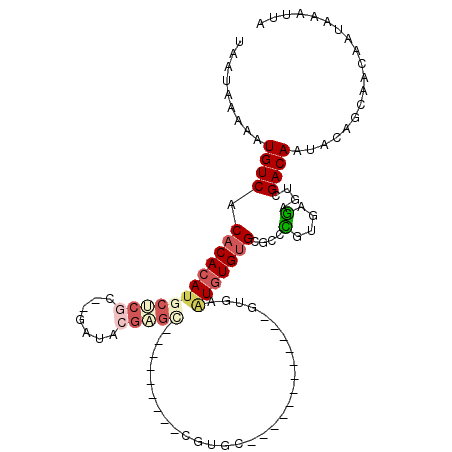
>2L_DroMel_CAF1 21950539 90 - 22407834 UAUUAAAAAUGUCACACACAUGCUCGC--GAUACGAGC--------CCGUGC------------GUGAAUGUGUGCGCCCGUGAGAGUCGACAAUACAGCAACAAUAAAUUA .........((((.(((((((..((((--(.((((...--------.)))))------------))))))))))).((.(....).)).))))................... ( -24.20) >DroPse_CAF1 15095 93 - 1 UAAUAAAAAUGUCACACACACACACACAU-------GG--------GUGCAUGGGUGUGU----GUAUAUCUGUGUGUGUGUGCGAGUCGACAAUACAGCAACAAUAAAUUA .........((((((.(.(((((((((((-------((--------((((((......))----))..))))))))))))))).).)).))))................... ( -30.70) >DroSec_CAF1 522 89 - 1 UAUUAAAAAUGUCACACACAUGCUCGC--GAUACGAGC---------CGAGC------------GUGAAUGUGUGCGCCCGUGAGAGUCGACAAUACAGCAACAAUAAAUUA .........((((((((((((((((((--.......).---------)))))------------)))..)))))..((.(....).)).))))................... ( -23.30) >DroSim_CAF1 3759 89 - 1 UAUUAAAAAUGUCACACACAUGCUCGC--GAUACGAGC---------CGGGC------------GUGAAUGUGUGCGCCCGUGAGAGUCGACAAUACAGCAACAAUAAAUUA .........((((((.(((..(((((.--....)))))---------.((((------------((........)))))))))...)).))))................... ( -25.40) >DroEre_CAF1 2490 98 - 1 UAAUAAAAAUGUCACACACAUGCUCGC--GAUACGUAUGUGUGCUCGCAUGU------------GUGAGUGUGUGUGCCCACGAGAGUCGACAAUACAGCAACAAUAAAUUA .........((((.(((((((((((((--(.....((((((....)))))))------------)))))))))))))...((....)).))))................... ( -29.40) >DroAna_CAF1 6677 102 - 1 UAAUAAAAAUGUCACACACAUGCACAG--AAUACGUGA--------CCGUAUGGGUCUGUAUCGGCGAGUGUGCGAGUGUGUGUAUGUCGACAAUACAGCAACAAUAAAUUA .........(((((((.((((((((.(--(.((((.((--------((.....)))))))))).(((....)))..)))))))).))).))))................... ( -27.60) >consensus UAAUAAAAAUGUCACACACAUGCUCGC__GAUACGAGC_________CGUGC____________GUGAAUGUGUGCGCCCGUGAGAGUCGACAAUACAGCAACAAUAAAUUA .........((((.((((((((((((.......)))))..............................)))))))....(....)....))))................... ( -7.82 = -8.88 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:55 2006