| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,947,016 – 21,947,120 |
| Length | 104 |
| Max. P | 0.995489 |

| Location | 21,947,016 – 21,947,120 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.56 |
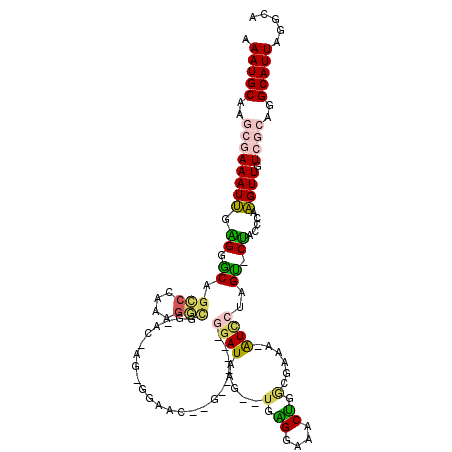
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -14.56 |
| Energy contribution | -13.98 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
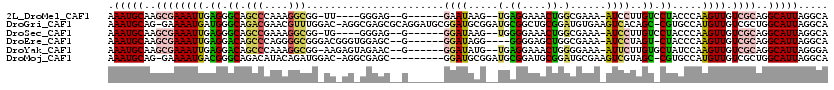
>2L_DroMel_CAF1 21947016 104 + 22407834 AAAUGCAAGCGAAAUUGAGGGCAGCCCAAAGGCGG-UU----GGGAG--G------GAAUAAG--UGAGGAAACUGGCGAAA-AUCCUUGUCCUACCCAAGUUGUCGCAGGCAUUAGGCA .(((((..((((..((((((((..(((((......-))----)))((--(------((....(--(.((....)).))....-.))))))))))...)))....))))..)))))..... ( -36.60) >DroGri_CAF1 10737 117 + 1 AAAUGCAG-GAAAAUGAUGGGCAGACGAACGUUUGGAC-AGGCGAGCGCAGGAUGCGGAUGCGGAUGCGGCUGCGGAUGUGAAGUCACAGC-CGUGCCAUGUUGUCGCUGGCAUUAGGCA .(((((..-........((..(((((....)))))..)-)(((((..(((...(((....)))..)))(((.((((.((((....)))).)-))))))......))))).)))))..... ( -40.60) >DroSec_CAF1 5643 104 + 1 AAAUGCAAGCGAAAUUGAGGGCAGCCGAAAGGCGG-UG----GGGAG--G------GGAUAAG--UGGGGAAACUGGCGAAA-AUCCUUGUCCUACCCAAGUUGUCGCAGGCAUUAGGCA .(((((..((((..(((......(((....)))((-((----(((..--(------((((..(--(.((....)).))....-)))))..))))))))))....))))..)))))..... ( -46.80) >DroEre_CAF1 1499 106 + 1 AAAUGCAAGCGAAAUUGAGGACAGCCCAGGGGCGGGACGGGUGGAGC--G------GGAUAGG----GGGGAGCUGGCGAAA-AUCCUAGU-CUACCCAAGUUGUCGCAGGCAUUAGGCA .(((((..((((((((..(..(.(((....))).)..)(((((((..--(------((((.(.----((....))..)....-)))))..)-)))))).)))).))))..)))))..... ( -36.50) >DroYak_CAF1 4383 108 + 1 AAAUGCAAGCGAAAUUGAGGACAGCCCAAAGGCGG-AAGAGUAGAAC--G------GGAUAUG--UGAGGAAACUGGGGAAA-AUUCUUGUGCUAUCCAAGUUGUCGCAGGCAUUAGGGA .(((((..((((((((..(((..(((....)))..-.....(((.((--(------(((..(.--(.((....)).).)...-..)))))).)))))).)))).))))..)))))..... ( -29.00) >DroMoj_CAF1 11237 108 + 1 AAAUGCAG-GAAAAUGACGGGCAGACAUACAGAUGGAC-AGGCGAGC---------GGAUGCGGAUGCGGAUGCGGAUGCGAAGUCGUAGC-CGUGCCAUGUUGUCGCUGGCAUUAGGCA ...(((..-...((((.(.(((.((((.(((..(((..-.(((..((---------((.((((..(((....)))..))))...)))).))-)...))))))))))))).)))))..))) ( -35.20) >consensus AAAUGCAAGCGAAAUUGAGGGCAGCCCAAAGGCGG_AC_AG_GGAAC__G______GGAUAAG__UGAGGAAACUGGCGAAA_AUCCUAGU_CUACCCAAGUUGUCGCAGGCAUUAGGCA .(((((..((((((((.((.((.(((....))).......................((((.....(.((....)).)......))))..)).)).....)))).))))..)))))..... (-14.56 = -13.98 + -0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:52 2006