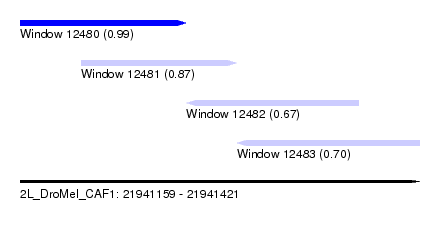
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,941,159 – 21,941,421 |
| Length | 262 |
| Max. P | 0.989731 |

| Location | 21,941,159 – 21,941,268 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.51 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -21.24 |
| Energy contribution | -22.52 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21941159 109 + 22407834 AUUUCUCACGAGAGAACCGUUUCCCGUACUCAGGCUAGUGAGCCUCAGAGUCAGAUGCAUCUGUAUCUGCCAGUUGUCAGGC------AUCACCAUCUCUAUUCUGAAUGCGAAC----- ..(((((....)))))........((((.((((..(((((((((((((...(((((((....)))))))....)))..))))------.))))......))..)))).))))...----- ( -30.70) >DroSec_CAF1 229384 109 + 1 AUUUCUCACGGGAGAACCGUUUCCCGUACUCAGGCUUGUCAGCCUCAGAGUCAGAUGCAUCCGUAUCUGCCAGUCGUCUGGC------AUCACCAUCUCUAUUCCGAAUGCGAAC----- ..(((....((((((....))))))(((.(((((((....))))).((((..((((((....))))))(((((....)))))------........)))).....)).)))))).----- ( -34.10) >DroSim_CAF1 174304 109 + 1 AUUUCUCACGGGAGAACCGUUUCCCGUACUCAGGCUUGUCGGCCUCAGAGUCAGAUGCAUCUGUAUCUGCCAGUCGUCUGGC------AUCACCAUCUCUAUUCCGAGUGCGAAC----- ..(((....((((((....))))))(((((((((((....))))).((((..((((((....))))))(((((....)))))------........)))).....))))))))).----- ( -40.40) >DroEre_CAF1 201841 94 + 1 AUUUCUCC--------------------UUCGGGCUUGUCAGCCUCAGAGGCAGUUGCAUCUGUAUCUGCCUGUCGUCAGGC------AUCACAAUCUUCAUUCUGAGUGCGAACGCGAA ......((--------------------....))...((..(((((((((((((.(((....))).)))))((((....)))------).............)))))).))..))..... ( -30.10) >DroYak_CAF1 193158 115 + 1 AUUUCUCACGAAAAACGCGUUUCACGUACUCAGGCUUGUCAGUCUCACAGGCAGUUGCAUUUGUAUCUGCCUGUCGUUAGGCGUCUUCAUCUCCAUUUUCAUUCUGAGUGGGAAC----- ..(((((((((..((.((((((.(((.....(((((....))))).((((((((.(((....))).))))))))))).)))))).))..)).......((.....))))))))).----- ( -34.60) >consensus AUUUCUCACGAGAGAACCGUUUCCCGUACUCAGGCUUGUCAGCCUCAGAGUCAGAUGCAUCUGUAUCUGCCAGUCGUCAGGC______AUCACCAUCUCUAUUCUGAGUGCGAAC_____ ........................((((((((((((....))))).((((..((((((....))))))(((((....)))))..............)))).....)))))))........ (-21.24 = -22.52 + 1.28)

| Location | 21,941,199 – 21,941,301 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -16.80 |
| Energy contribution | -18.16 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21941199 102 + 22407834 AGCCUCAGAGUCAGAUGCAUCUGUAUCUGCCAGUUGUCAGGC------AUCACCAUCUCUAUUCUGAAUGCGAAC------------GCGAAUCUACGCCAUCUCGCGUCACCCACUUAA .((.(((((((.((((((....))))))(((........)))------............)))))))..))((.(------------((((............))))))).......... ( -25.80) >DroSec_CAF1 229424 81 + 1 AGCCUCAGAGUCAGAUGCAUCCGUAUCUGCCAGUCGUCUGGC------AUCACCAUCUCUAUUCCGAAUGCGAAC------------GCGAAUCCCCGG--------------------- ..((..((((..((((((....))))))(((((....)))))------........))))........(((....------------))).......))--------------------- ( -19.40) >DroSim_CAF1 174344 102 + 1 GGCCUCAGAGUCAGAUGCAUCUGUAUCUGCCAGUCGUCUGGC------AUCACCAUCUCUAUUCCGAGUGCGAAC------------GCGAAUCCCCGCCAUCUCGCGUCACCCACUUAA ((....((((..((((((....))))))(((((....)))))------........))))...))(((((.(.((------------((((............))))))..).))))).. ( -30.00) >DroEre_CAF1 201861 114 + 1 AGCCUCAGAGGCAGUUGCAUCUGUAUCUGCCUGUCGUCAGGC------AUCACAAUCUUCAUUCUGAGUGCGAACGCGAACGCGAACGCGAAUCCCCGCCAUCUCGCGUCACCCACUUAA .(((((((((((((.(((....))).)))))((((....)))------).............)))))).))......((.(((((..(((......)))....))))))).......... ( -37.30) >DroYak_CAF1 193198 108 + 1 AGUCUCACAGGCAGUUGCAUUUGUAUCUGCCUGUCGUUAGGCGUCUUCAUCUCCAUUUUCAUUCUGAGUGGGAAC------------GCGAAACUCCGCCAUCUCGCGUCAGCCACUAAA ......((((((((.(((....))).)))))))).....(((.......((.((((((.......))))))))((------------((((............))))))..)))...... ( -34.40) >consensus AGCCUCAGAGUCAGAUGCAUCUGUAUCUGCCAGUCGUCAGGC______AUCACCAUCUCUAUUCUGAGUGCGAAC____________GCGAAUCCCCGCCAUCUCGCGUCACCCACUUAA .(((((((((..((((((....))))))(((((....)))))...................))))))).))................(((......)))..................... (-16.80 = -18.16 + 1.36)


| Location | 21,941,268 – 21,941,381 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.56 |
| Mean single sequence MFE | -43.31 |
| Consensus MFE | -40.29 |
| Energy contribution | -40.92 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21941268 113 - 22407834 GGACAGGUCUCCGCCGUAUCUGUCGUGCCAUCUGAAUCUCAUUGCCAUUUGAGCAUCCGGCCGUAGGAGCUACUGGCAAAUUAAGUGGGUGACGCGAGAUGGCGUAGAUUCGC------- ...(..((((.((((((.....(((((.((((((.......((((((....(((.(((.......))))))..))))))......)))))).))))).)))))).))))..).------- ( -39.52) >DroSim_CAF1 174413 113 - 1 GGACAGGUCUUCGCCGUAUCUGUCGUGCCAUCUGAAUCUCAUUGCCAUUUGAGCAUCCGGCCGUAGGAGCUACUGGCAAAUUAAGUGGGUGACGCGAGAUGGCGGGGAUUCGC------- ...(..(((((((((((.....(((((.((((((.......((((((....(((.(((.......))))))..))))))......)))))).))))).)))))))))))..).------- ( -42.62) >DroEre_CAF1 201935 119 - 1 GGACAGGUCUCCGCCGUAUUCGUCGUGCCAUCUGAAUCUCAUUGCCAUUUGAGCAUCCGGCCGUAG-AGCUACUGGCAAAUUAAGUGGGUGACGCGAGAUGGCGGGGAUUCGCGUUCGCG ((((.((((((((((((.....(((((.((((((.......((((((....(((.((........)-))))..))))))......)))))).))))).)))))))))).))..))))... ( -43.72) >DroYak_CAF1 193273 113 - 1 GGACAGGUCUCCGCCGUAUUUGUCGUGCCAUCUGAAUCUCAUUGCCAUUUGAGCAUCCGGUUGUAGGAGCUACUGGCAAGUUUAGUGGCUGACGCGAGAUGGCGGAGUUUCGC------- .....((.(((((((((...(((((.((((.((((((....((((((....(((.(((.......))))))..)))))))))))))))))))))....)))))))))..))..------- ( -47.40) >consensus GGACAGGUCUCCGCCGUAUCUGUCGUGCCAUCUGAAUCUCAUUGCCAUUUGAGCAUCCGGCCGUAGGAGCUACUGGCAAAUUAAGUGGGUGACGCGAGAUGGCGGAGAUUCGC_______ ...(..(((((((((((.....(((((.((((((.......((((((....(((.(((.......))))))..))))))......)))))).))))).)))))))))))..)........ (-40.29 = -40.92 + 0.63)


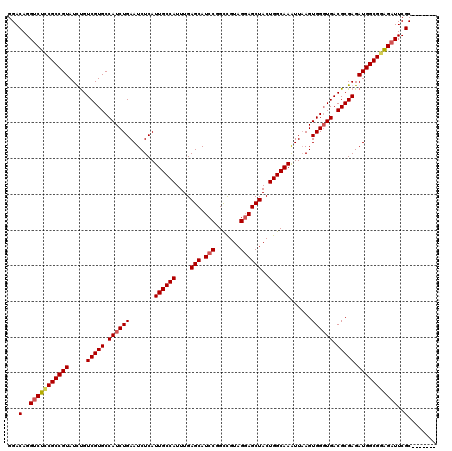
| Location | 21,941,301 – 21,941,421 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -40.77 |
| Consensus MFE | -39.97 |
| Energy contribution | -40.10 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21941301 120 - 22407834 UUUGUCAGAAUGUAAACUGAAUGGGAAUAGGAAGACGGCGGGACAGGUCUCCGCCGUAUCUGUCGUGCCAUCUGAAUCUCAUUGCCAUUUGAGCAUCCGGCCGUAGGAGCUACUGGCAAA .(((((((..........((((((.(((..(((((.((((.((((((((......).))))))).)))).)))...))..))).)))))).(((.(((.......)))))).))))))). ( -44.90) >DroSim_CAF1 174446 120 - 1 UUUGUCAGGAUGUAAACUGAAUGGGAAUAGGAAGACGGCGGGACAGGUCUUCGCCGUAUCUGUCGUGCCAUCUGAAUCUCAUUGCCAUUUGAGCAUCCGGCCGUAGGAGCUACUGGCAAA .(((((((..........((((((.(((..(((((.((((.((((((((......).))))))).)))).)))...))..))).)))))).(((.(((.......)))))).))))))). ( -45.10) >DroEre_CAF1 201975 119 - 1 UUUGUCAGGACGUAAACUGAAUGGGAAUAGGAAGACGGCGGGACAGGUCUCCGCCGUAUUCGUCGUGCCAUCUGAAUCUCAUUGCCAUUUGAGCAUCCGGCCGUAG-AGCUACUGGCAAA ....((((........))))((((.(..(.(((.(((((((((....)).))))))).))).)..).))))..........((((((....(((.((........)-))))..)))))). ( -37.20) >DroYak_CAF1 193306 119 - 1 UUUGUCGGAAUGUGAGCUGAAUGGGAAUAGGAAGACGG-GGGACAGGUCUCCGCCGUAUUUGUCGUGCCAUCUGAAUCUCAUUGCCAUUUGAGCAUCCGGUUGUAGGAGCUACUGGCAAG .(((((((((((.((..(..((((.(..(.(((.((((-.(((......))).)))).))).)..).))))..)..)).))))........(((.(((.......)))))).))))))). ( -35.90) >consensus UUUGUCAGAAUGUAAACUGAAUGGGAAUAGGAAGACGGCGGGACAGGUCUCCGCCGUAUCUGUCGUGCCAUCUGAAUCUCAUUGCCAUUUGAGCAUCCGGCCGUAGGAGCUACUGGCAAA ((((((((..........((((((.(((..(((((.((((.((((((((......).))))))).)))).)))...))..))).)))))).(((.(((.......)))))).)))))))) (-39.97 = -40.10 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:51 2006