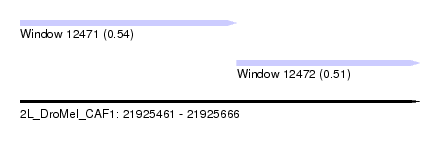
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,925,461 – 21,925,666 |
| Length | 205 |
| Max. P | 0.541469 |

| Location | 21,925,461 – 21,925,572 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -24.67 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
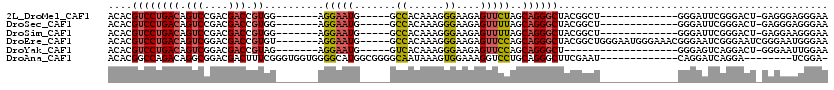
>2L_DroMel_CAF1 21925461 111 + 22407834 AAAAAAGAUGCCU---------GGAAGCCAAAGAGACAGCACUGGUAAAUGCACAACACGUGAACCAGGGCCACGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUC .............---------((..((((((..(((.((.(((((.....(((.....))).))))).))...((((((..(((.........)))..)))))).))).))))))..)) ( -31.00) >DroSec_CAF1 212992 111 + 1 AAAAAAGAUGCCU---------GGAAGCCAAAGAGACAGCACUGGUAAAUGCACAACACGUGAACCAGGGCCACGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUC .............---------((..((((((..(((.((.(((((.....(((.....))).))))).))...((((((..(((.........)))..)))))).))).))))))..)) ( -31.00) >DroSim_CAF1 157327 111 + 1 AAAAAAGAUGCCU---------GGAAGCCAAAGAGACAGCACUGGUAAAUGCACAACACGUGAACCAGGGCCACGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUC .............---------((..((((((..(((.((.(((((.....(((.....))).))))).))...((((((..(((.........)))..)))))).))).))))))..)) ( -31.00) >DroEre_CAF1 180031 120 + 1 AAAAAAUAUGCCGGAGCUACGAGGAAGCCAAAGAUACAGCACUGGUAAAUGCACAACAUGUGAACCAGGGCCACGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUC ...........((......)).((..(((((((((...((.(((((.....(((.....))).))))).))...((((((..(((.........)))..)))))).))).))))))..)) ( -30.20) >DroYak_CAF1 175508 120 + 1 AAAAGCGAUGCCGGGGUUACGCGGAAGCCAAAGAAACAGCACUGGUAAAUGCACAACAUGUGAACCAGGGCCACGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUC ....(((...(((.(....).)))..((((((...((.((.(((((.....(((.....))).))))).))...((((((..(((.........)))..)))))).))..))))))))). ( -34.50) >DroAna_CAF1 155327 117 + 1 CAA---UUUAAAGCAGCCAUAAAACAGCUAGAAGUACAGCACUUGGAGAGGCCCUACACGUGAGCCAGGGCCACGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUC ...---......((.((((....((..((..((((.....))))..)).((((((...(....)..))))))..((((((..(((.........)))..)))))).))....)))).)). ( -31.90) >consensus AAAAAAGAUGCCG_________GGAAGCCAAAGAGACAGCACUGGUAAAUGCACAACACGUGAACCAGGGCCACGUCAGUCAGCAAUAAAUAAAUGCAAGCUGGCAGUCAUUUGGUCGUC ......................((..((((((...((.((.(((((.....(((.....))).))))).))...((((((..(((.........)))..)))))).))..))))))..)) (-24.67 = -24.92 + 0.25)

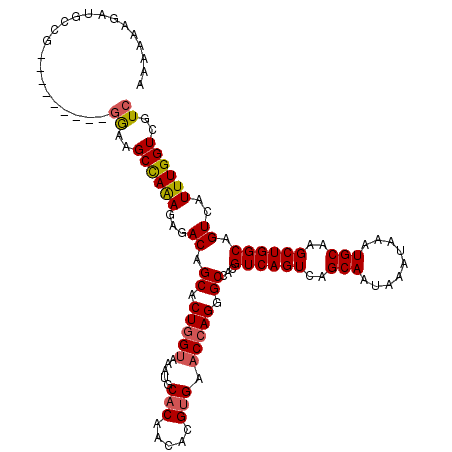
| Location | 21,925,572 – 21,925,666 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -12.24 |
| Energy contribution | -12.35 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.37 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21925572 94 + 22407834 ACACGUCCUGACAGUCCGACGACCGUGG-------AGGAAUG-----GCCACAAAGGGAAGAGUUCUAGCAGGGCUACGGCU-------------GGGAUUCGGGACU-GAGGGAGGGAA ...(.((((..((((((.......((((-------.......-----.))))........((((((((((.(.....).)))-------------))))))).)))))-).)))).)... ( -37.40) >DroSec_CAF1 213103 94 + 1 ACACGUCCUGACAGUCCGACGACCGUGG-------AGGAAUG-----GCCACAAAGGGAAGAGUUUUAGCAGGGCUACGGCU-------------GGGAUUCGGGACU-GAGGGAGGGAA ...(.((((..((((((.......((((-------.......-----.))))........((((((((((.(.....).)))-------------))))))).)))))-).)))).)... ( -34.80) >DroSim_CAF1 157438 94 + 1 ACACGUCCUGACAGUCCGACGACCGUGG-------AGGAAUG-----GCCACAAAGGGAAGAGUUUUAGCAGGGCUACGGCU-------------GGGAUUCGGGACU-GAGGAAGGGAA .(...((((..((((((.......((((-------.......-----.))))........((((((((((.(.....).)))-------------))))))).)))))-)))))...).. ( -31.60) >DroEre_CAF1 180151 108 + 1 ACACGUCCUGACAGUCGGACGACCGUGU-------AGGAAUG-----GCCACAAAGGGAAGAGUUCCAGCAGGGCUACGGCUGGGAAUGGGAAACGGGAAUCGGGAAUCGGGAAUGGGAA .(...((((((((((((...(.((.(((-------.(((((.-----.((......))....))))).))).)))..)))))).((.(.(....).)...)).....))))))...)... ( -34.90) >DroYak_CAF1 175628 88 + 1 ACACGUCCUGACAGUCGGACGACCGUAG-------AGGAAUG-----GUCACAAAGGGAAGAGUUCCAGCAGGGCU-------------------GGGAGUCAGGACU-GGGAAUUGGAA .((.((((((((........((((((..-------....)))-----))).............(((((((...)))-------------------)))))))))))))-).......... ( -32.50) >DroAna_CAF1 155444 98 + 1 ACACGGCCAGACAGGCGGACGACUUUCGGGUGGUGGGGCAUGGCGGGGCAAUAAAGUGGAAAGGUCCUGCAGGGCUUCGAAU-------------CAGGAUCAGGA--------UCGGA- ...(.(((.....))).).((((((((.(((..((((((...(((((((..............)))))))...)))))).))-------------)..))..))).--------)))..- ( -29.94) >consensus ACACGUCCUGACAGUCCGACGACCGUGG_______AGGAAUG_____GCCACAAAGGGAAGAGUUCCAGCAGGGCUACGGCU_____________GGGAUUCGGGACU_GAGGAAGGGAA ....((((((((.(((....))).))..........(((((.......((......))....)))))..))))))............................................. (-12.24 = -12.35 + 0.11)
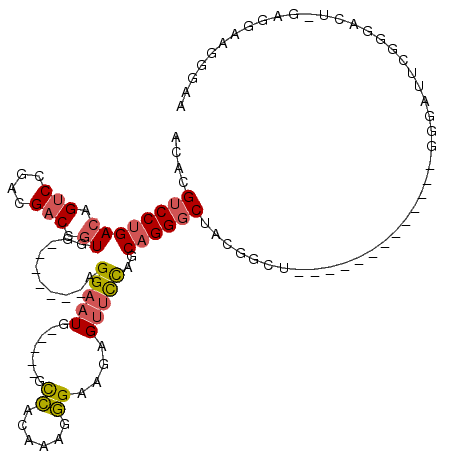
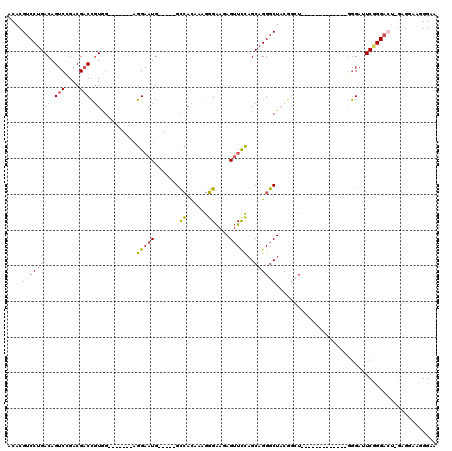
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:41 2006