
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,924,719 – 21,924,838 |
| Length | 119 |
| Max. P | 0.635167 |

| Location | 21,924,719 – 21,924,838 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -19.71 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
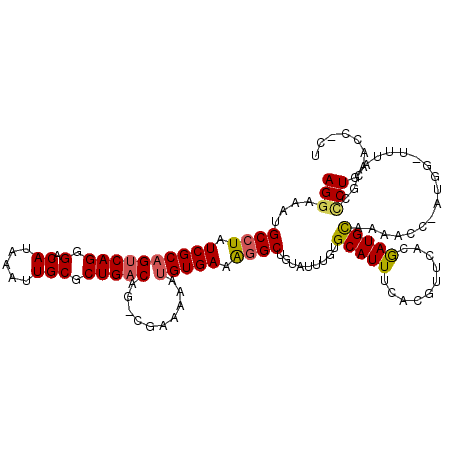
>2L_DroMel_CAF1 21924719 119 + 22407834 AGGAAAUGCCUAUCGCAGUCAGGGACAUAAAUUGCGCUGACAG-CGAAAAAUGUGAAAGGCUGUAUUUGUGCAUUUAACGUUCACGAUGCAAAACCCAUGGCUUCACGCCCUGAACCACU (((.........((((.(((((.(.((.....))).))))).)-))).....((((..((((((.....((((((..........))))))......))))))))))..)))........ ( -31.30) >DroSec_CAF1 212222 113 + 1 AGGAAAUGCCUAUCGCAGCCAGGGACAUAAAUUGCGCUGACAGGAGGUGUUUGUGAAGGGCUGUAU-UGUGCAUUUCACGUUCACAAUGUAAAACC------UUUACGCCCUGAACCCCU ..(((((((.((..((((((.....(((((...(((((.......))))))))))...))))))..-)).)))))))..(((((...(((((....------.)))))...))))).... ( -32.30) >DroSim_CAF1 156518 114 + 1 AGGAAAUGCCUAUCGCAGUCAGGGACAUAAAUUGCGCUGACAGGCUGUGUUUGUGAAAGGCUGUAUUUGUGCAUUUCACGUUCACAAUGUAAAACC------UUUACGCCCUGAACCCCU (((...(((.....)))(((((.(.((.....))).))))).(((((((..(((((((.((.........)).)))))))..))))..((((....------.))))))).......))) ( -32.30) >DroEre_CAF1 178509 108 + 1 AGGAAAUGCCUAUCGCAGUCAGGGACAUAAAUUGCGCUGACAG-CGAAAAAUGUGAAAGGCUGUAUUUGUGCAUUUCACGUUCACGAUGC-------AGGGUUAUACGCCCUGCG----U (((.....))).((((.(((((.(.((.....))).))))).)-)))..(((((((((.((.........)).)))))))))....((((-------(((((.....))))))))----) ( -40.60) >DroYak_CAF1 174154 115 + 1 AGGAAAUGCCUAUCGCAGUCAGGGACAUAAAUUGCACUGACAG-AGAAAAAUGUGAAAGGCGGUAUUUGUGCAUUUCACAUUCACGAUGCUAAACCCAUGACUAUACACACUGCG----C .((....((((.((((((((((.(.((.....))).)))))..-.......))))).))))((((((.(((.((.....)).)))))))))....))..................----. ( -24.40) >DroAna_CAF1 154724 116 + 1 AGGAAAUGCCUAUCGCUGUCAGGGACAUAAAUUGCGCUGACAG-CGAAAAAUGUGAAAAGCCACAUUUGUGCAUUUCGCGUUGACGAUGCAAAACCCUUGGA---CCUCUCUGGGCCUUU ..(((((((...((((((((((.(.((.....))).)))))))-))).(((((((......)))))))..)))))))(((((...)))))....(((..((.---...))..)))..... ( -43.80) >consensus AGGAAAUGCCUAUCGCAGUCAGGGACAUAAAUUGCGCUGACAG_CGAAAAAUGUGAAAGGCUGUAUUUGUGCAUUUCACGUUCACGAUGCAAAACC_AUGG_UUUACGCCCUGAACC_CU (((....((((.((((((((((.(.((.....))).)))))..........))))).)))).........(((((..........)))))...................)))........ (-19.71 = -19.68 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:38 2006