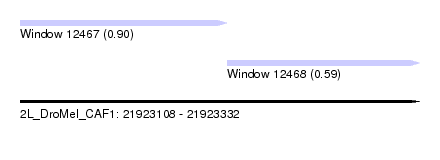
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,923,108 – 21,923,332 |
| Length | 224 |
| Max. P | 0.897720 |

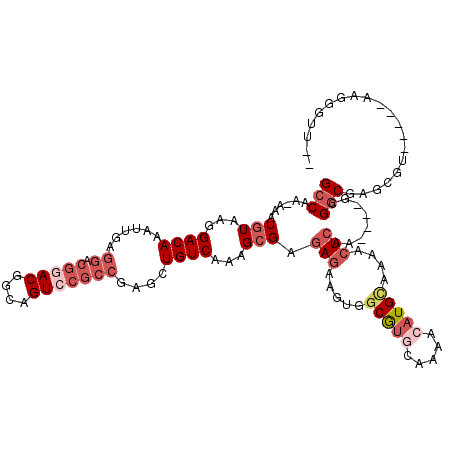
| Location | 21,923,108 – 21,923,224 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -14.52 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21923108 116 + 22407834 CACGCCAAUAAAUUUCAAAUGUGUGGCGCGGAUUGUCCAACGCCUCGCACUGCUCUUUCUCAAAGCAGGCGAAAUGAAAAAAAAAAAAAAA----UUGAAAAGGAAGACAAACCGGCAAC ...(((......((((((......((((.((.....))..))))((((.(((((.........)))))))))...................----)))))).((........)))))... ( -30.00) >DroSec_CAF1 210489 108 + 1 CACGCCAAUAAAUUUCCAAUGUGUGGCGCGGAUUGUCCAACGCCUCGCACUGCUCUUUCCCAAAGCAGGCGAAAUGAAA--------AAAA----UUGAAAAGGAAGACAAACCGGCAAC ...(((......(((((.......((((.((.....))..))))((((.(((((.........))))))))).......--------....----.......))))).......)))... ( -28.62) >DroSim_CAF1 155067 108 + 1 CACGCCAAUAAAUUUCCAAUGUGUGGCGCGGAUUGUCCAACGCCUCGCACUGCUCUUUCCCAAAGCAGGCGAAAUGAAA--------AAAA----UUGAAAAGGAAGACAAACCGGCAAC ...(((......(((((.......((((.((.....))..))))((((.(((((.........))))))))).......--------....----.......))))).......)))... ( -28.62) >DroEre_CAF1 176702 105 + 1 CACGCCAAUAAAAUUCAAAUGUGUGGCACGGAUUGCCCAAUGCCUCACACAA---UGCCUCAAGGCAGUCGAAAUGAAA--------AAAU----UUGAAAAAGAAGACAAACCGGCAAC ...(((.......((((((((((.((((.((.....))..)))).)))....---((((....))))............--------..))----)))))......(......))))... ( -25.60) >DroYak_CAF1 172564 105 + 1 CACGCCAAUACAAUUCAAAUGUGUGGUGUGGAUUGUCCAAUGCCUCGCACCA---UGCCUCAAGGCAGCCGAAAUGAAA--------AAAA----AUGAAAAGGAAGACAAACUGGCAAC ...((((...((.(((...((((.(((((((.....)).))))).))))...---((((....))))...))).))...--------....----.......(.....)....))))... ( -25.10) >DroAna_CAF1 153498 99 + 1 CGAGCUAAUAAAAUUCAAAUGUGUGGCGCGGACUGUCCGCUGCCUGAUCUCGCAAAUUGUCAAAGCGGGCGAAAUGAGA--------AAAAAACCCUGAG--------CAAAAG-----G ...(((..................(((((((.....)))).)))...(((((....(((((......)))))..)))))--------...........))--------).....-----. ( -22.70) >consensus CACGCCAAUAAAAUUCAAAUGUGUGGCGCGGAUUGUCCAACGCCUCGCACUGCUCUUUCUCAAAGCAGGCGAAAUGAAA________AAAA____UUGAAAAGGAAGACAAACCGGCAAC ...(((.............((((.((((.((.....))..)))).))))((((...........))))..............................................)))... (-14.52 = -15.17 + 0.64)

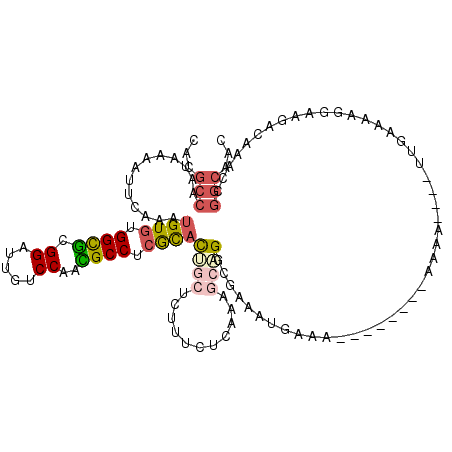
| Location | 21,923,224 – 21,923,332 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -18.26 |
| Energy contribution | -20.43 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21923224 108 + 22407834 GCCAA-AAAUGUAAGGACAAAUUGAGGACGGACGACAGUCCGCCGAGCUGUCAAAGCGAGAGAAGUGGCGUGCAAAACAUGCAAAACUCAA----GCGGCGAGCGU-----AAGGAUU-- (((..-.........((((......((.(((((....)))))))....))))...((..(((.....(((((.....)))))....)))..----)))))...(..-----..)....-- ( -32.50) >DroSec_CAF1 210597 108 + 1 GCCAA-AAAUGUAAGGACAAAUUGAGGACGGACGGCAGUCCGCCGAGCUGUCAAAGCGAAAGAAGUGGCGUGCAAAACAUGCAAAACUCAA----GCGGCGAGCGU-----AAGGGUU-- (((..-..((((.............((.(((((....)))))))..(((((((...(....)...))(((((.....))))).........----)))))..))))-----...))).-- ( -30.70) >DroSim_CAF1 155175 108 + 1 GCCAA-AAAUGUAAGGACAAAUUGAGGACGGACGGCAGUCCGCCGAGCUGUCAAAGCGAGAGAAGUGGCGUGCAAAACAUGCAAAACUCAA----GCGGCGAGCGU-----AAGGGUU-- (((..-.........((((......((.(((((....)))))))....))))...((..(((.....(((((.....)))))....)))..----)))))...(..-----..)....-- ( -32.50) >DroEre_CAF1 176807 109 + 1 GCCAAAAAAUGUAAGGACAAAUUGAGGACGGACGGCAGUCCGCCGAGCUGUCAAAGCGAGAGAAGUGGCGUGCGAAACAUGCAAAACUCGA----GCGGCAGGCGU-----AAAGGGU-- (((............((((......((.(((((....)))))))....))))...((..(((.....(((((.....)))))....)))..----)))))......-----.......-- ( -33.40) >DroYak_CAF1 172669 107 + 1 GCCAA-AAAUGUAAGGACAAAUUGAGGACGGACGACAGUACGCCGAGCUGUCAAAGCGAGAGAAGUGGCGUGCGAAAAAUGUAAAACUCAA----GCGGCGAGUUU------AGGAGU-- (((..-...(((....)))..(((((.(((.......((((((((..((.((.....)).))...))))))))......)))....)))))----..)))......------......-- ( -30.02) >DroAna_CAF1 153597 105 + 1 GCCAAACAAUAUAA-GACAAAGUGAGGACGG--------------AGCUGUCGAGGCGAGAGAAGUGCCACAGAAAACAUGCAAAAAUCAGUCCAGAGUCCAGAGAUGGAGAUGGGUUGU ..............-......(((.(.((..--------------..((.(((...))).))..)).)))).................((..(((...((((....))))..)))..)). ( -20.70) >consensus GCCAA_AAAUGUAAGGACAAAUUGAGGACGGACGGCAGUCCGCCGAGCUGUCAAAGCGAGAGAAGUGGCGUGCAAAACAUGCAAAACUCAA____GCGGCGAGCGU_____AAGGGUU__ (((......(((...((((......((.(((((....)))))))....))))...))).(((.....(((((.....)))))....)))........))).................... (-18.26 = -20.43 + 2.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:37 2006