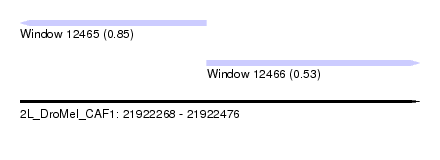
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,922,268 – 21,922,476 |
| Length | 208 |
| Max. P | 0.852471 |

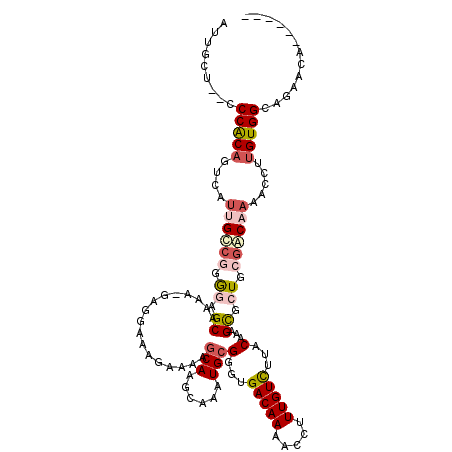
| Location | 21,922,268 – 21,922,365 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 90.89 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -28.14 |
| Energy contribution | -28.33 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21922268 97 - 22407834 CAGCAAUGUAGGCGUUGAAAAGCCACUGGUACCUGCCACCGGGGCUGGCGCUGUUGGAAUGGGCAGCAAAUUUGCAAUGUGUACUUCACGGAUGUAU .......(((.((((((...((((.(((((.......)))))))))...((((((......)))))).......)))))).)))............. ( -29.80) >DroSec_CAF1 209691 97 - 1 CUGCAAUGUAGGCGUUGCAAAGCCACUGUUACCUGCCACCGGGGCUGGCGCUGUUGGAAUGGGCAGCAAAUUUGCAACGUGUACUUUACGGAUGUAU (((.((.(((.((((((((((((((......((((....))))..))))((((((......))))))...)))))))))).))).)).)))...... ( -36.80) >DroSim_CAF1 154246 97 - 1 CUGCAAUGUAGGCGUUGCAAAGCCACUGGUACCUGCCACCGGGGCUGGCGCUGUUGGAAUGGGCAGCAAAUUUGCAACGUGUACUUUACGGAUGUAU (((.((.(((.(((((((((((((.(((((.......)))))))))...((((((......))))))....))))))))).))).)).)))...... ( -39.30) >DroYak_CAF1 171274 97 - 1 CUGCAAUGUAGGCGUUGUAAAGCCUCCGGUUCCUGCCACCGGGACCGGUGGUGUUGGAAUGGGCAGUAUAUUUGUAUCGUGUACUUUAGGGAUGUAU .(((((((....)))))))..((((...(((((.(((((((....)))))))...))))))))).((((((((.((..(....)..)).)))))))) ( -36.00) >consensus CUGCAAUGUAGGCGUUGCAAAGCCACUGGUACCUGCCACCGGGGCUGGCGCUGUUGGAAUGGGCAGCAAAUUUGCAACGUGUACUUUACGGAUGUAU .......(((.(((((((((((((((((((.......)))))...))))((((((......))))))...)))))))))).)))............. (-28.14 = -28.33 + 0.19)

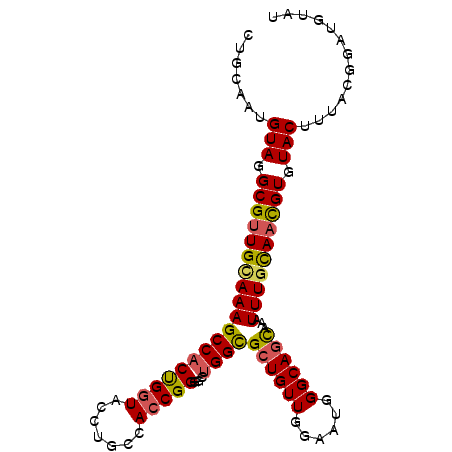
| Location | 21,922,365 – 21,922,476 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -17.47 |
| Energy contribution | -18.58 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21922365 111 + 22407834 AUUGCU--CCCGCAGUCAAUGCCGGGGAGCAAAA-GAGGAAAGAAAAGCAAGCACAUGCGGGUGACAAAACCUUUGUCUUACAAAGCGGUGCGGCAAAACCUUGUGGCAGAACG------ .(((((--((((((.....)))..))))))))..-............((..((((.(((.(..(((((.....)))))...)...))))))).((((....)))).))......------ ( -33.70) >DroSec_CAF1 209788 111 + 1 AUUGCU--GCCACAGUCAUUGCCGGGGAGCCAAA-GAGGAAAGAAAAGCAAGCACAUGCGGGUGACAAAACCUUUGUCUUACAAAGCGCUGCGACAAAACCUUGUGGCAGAACA------ ....((--(((((((...(((.((.((.((....-.((((.......(((......)))((((......))))...)))).....)).)).)).)))....)))))))))....------ ( -31.40) >DroSim_CAF1 154343 111 + 1 AUUGCU--GCCACAGUCAUUGCCGGGGAGCCAAA-GAGGAAAGAAAAGCAAGCACAUGCGGGUGACAAAACCUUUGUCUUACAAAGCGCUGCGACAAAACCUUGUGGCAGAACA------ ....((--(((((((...(((.((.((.((....-.((((.......(((......)))((((......))))...)))).....)).)).)).)))....)))))))))....------ ( -31.40) >DroEre_CAF1 175842 111 + 1 AUUGCU--CCCACAUUCAUGGUCGGGGAGCAAAAGGAAGAAAGAAAAGCA-GAAAAUGCGGUUGACAAAACAUUUGUCUUACAAAGCGCUGCGACAAAACCUCGUGGUAGAACA------ .(((((--(((.((....))....))))))))...............(((-(....(((.((.(((((.....)))))..))...)))))))......(((....)))......------ ( -28.40) >DroYak_CAF1 171371 112 + 1 AUUGCU--CCCACAUUCGUGGUCGGAGAGCAAAAAGAAGAAAUAAAAGCAAGCAAAUGCGGUUGACAAAACCUUUGUCUUACAAAGUGCUGGGACAAAGCCUUGUGGUAGAACA------ .(((((--(((((....)))).....))))))...............((((((......((((.....))))(((((((((........)))))))))).))))).........------ ( -26.80) >DroAna_CAF1 153023 113 + 1 CAUGUCUCCCCGCAUUCAUUGUCGAGUUGCA-------GAAACAAAAACAAGCGAAUGCGGGUGACAAAAGUUUUGUUGUACAAAGUGCUCGAACAAAACCUUGUGGUAGGUGAGCACAG ..((((..(((((((((.((((...(((...-------..)))....))))..))))))))).)))).....((((.....))))(((((((......(((....)))...))))))).. ( -37.70) >consensus AUUGCU__CCCACAGUCAUUGCCGGGGAGCAAAA_GAGGAAAGAAAAGCAAGCAAAUGCGGGUGACAAAACCUUUGUCUUACAAAGCGCUGCGACAAAACCUUGUGGCAGAACA______ .........(((((....((((((.((.((.................(((......)))(...(((((.....)))))...)...)).)).)))))).....)))))............. (-17.47 = -18.58 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:35 2006