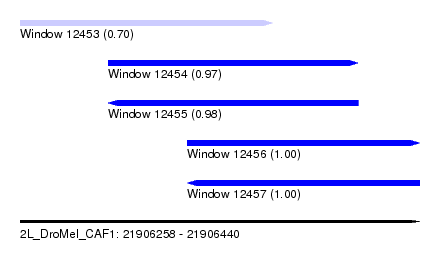
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,906,258 – 21,906,440 |
| Length | 182 |
| Max. P | 0.999926 |

| Location | 21,906,258 – 21,906,373 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.66 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21906258 115 + 22407834 ACUCAUUCAAUAGUUUUGAAAAUUGCGAAUGUCACUUACAACCCAACAACAAGAGCCACUGCCUUGCACUUACAGC----UCCCUCUUUUCUCAGGCACAUGUGUCUGUGAGUGAGCAU ...(((((..(((((.....))))).)))))..................((((.((....))))))........((----((.(((......((((((....)))))).))).)))).. ( -23.50) >DroSec_CAF1 194894 119 + 1 ACUCAUUCAAUAGUUUUGAAAAUUGCGAAUGUCACUUACAACCCAACAACAAGAGCCACUGCCUUGCACUUACAGGCACAUGCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUGAGCAU .....(((((.....)))))...(((.......................((((.((....))))))(((((((((((((((((((........)))))..)))))))))))))).))). ( -38.00) >DroSim_CAF1 138925 115 + 1 ACUCAUUCAAUAGUUUUGAAAAUUGCGAAUGUCACUUACAACCCAACAACAAGAGCCACUGCCUUGCACUUACAGC----UCCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUGAGCAU ...(((((..(((((.....))))).)))))..................((((.((....))))))........((----((.(((......((((((....)))))).))).)))).. ( -23.50) >DroEre_CAF1 158462 115 + 1 ACUCAUUCAAUAGUUUUGAAAAUUGCGAAUGUCACUUACAACCCAACAACAAGAGCCACUGCCUUGCACUUACAGC----UCCCUCUUUCCUCGGGCACAUGUGUCUGUGAGUGAACAU .....(((((.....)))))........((((((((((((.........((((.((....))))))....((((..----.(((.........)))....))))..)))))))).)))) ( -24.00) >DroYak_CAF1 155048 115 + 1 ACUCAUUCAAUAGUUUUGAAAAUUACGAAUGUCACUUACAUCCCAACAACAAGAACCACUGCCUUGCACUUACAGC----UUCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUAAGCAU ...(((((..(((((.....))))).)))))..................((((.........))))........((----((.(((......((((((....)))))).))).)))).. ( -18.70) >consensus ACUCAUUCAAUAGUUUUGAAAAUUGCGAAUGUCACUUACAACCCAACAACAAGAGCCACUGCCUUGCACUUACAGC____UCCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUGAGCAU .....(((((.....)))))........(((((((((((..........((((.((....))))))...........................(((((....))))))))))))).))) (-20.42 = -20.66 + 0.24)

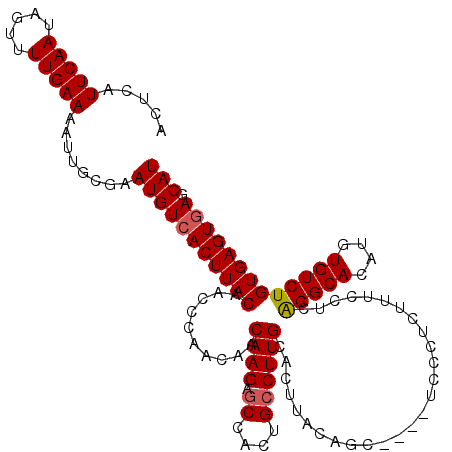
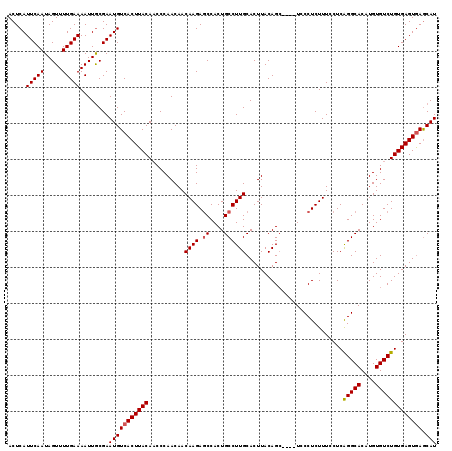
| Location | 21,906,298 – 21,906,412 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 94.91 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -32.40 |
| Energy contribution | -32.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21906298 114 + 22407834 ACCCAACAACAAGAGCCACUGCCUUGCACUUACAGC----UCCCUCUUUUCUCAGGCACAUGUGUCUGUGAGUGAGCAUCCUUGCCAAGGAUGCGGACUGAGACAAUGUUGCCGGAGA- .(((((((.((((.((....))))))......(((.----(((.((..(((.((((((....)))))).))).))(((((((.....)))))))))))))......)))))..))...- ( -37.20) >DroSec_CAF1 194934 118 + 1 ACCCAACAACAAGAGCCACUGCCUUGCACUUACAGGCACAUGCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUGAGCAUCCUUGCCAAGGAUGCGGACUGAGACAAUGUUGCCGGAGA- .(((((((.((((.((....))))))(((((((((((((((((((........)))))..)))))))))))))).(((((((.....)))))))............)))))..))...- ( -48.50) >DroSim_CAF1 138965 114 + 1 ACCCAACAACAAGAGCCACUGCCUUGCACUUACAGC----UCCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUGAGCAUCCUUGCCAGGGAUGCGGACUGAGACAAUGUUGCCGGAGA- .(((((((.((((.((....))))))......(((.----(((.((.(((..((((((....)))))).))).))(((((((.....)))))))))))))......)))))..))...- ( -36.90) >DroEre_CAF1 158502 115 + 1 ACCCAACAACAAGAGCCACUGCCUUGCACUUACAGC----UCCCUCUUUCCUCGGGCACAUGUGUCUGUGAGUGAACAUCCUUGCCAAGGAUGCGGACUGAGACAAUGUUGCCGGAGAC .(((((((.((((.((....))))))......(((.----(((.((.(((..((((((....)))))).))).)).((((((.....)))))).))))))......)))))..)).... ( -31.50) >DroYak_CAF1 155088 114 + 1 UCCCAACAACAAGAACCACUGCCUUGCACUUACAGC----UUCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUAAGCAUCCUUGCCAAGGAUGCGGACUGAGACAAUGUUGCCGGAGA- ((((((((.((((.........))))..........----...(((..(((.((((((....)))))).......(((((((.....))))))))))..)))....)))))..)))..- ( -33.10) >consensus ACCCAACAACAAGAGCCACUGCCUUGCACUUACAGC____UCCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUGAGCAUCCUUGCCAAGGAUGCGGACUGAGACAAUGUUGCCGGAGA_ .........((((.((....))))))........................(((.(((((((.(((((...(((..(((((((.....)))))))..))).)))))))).)))).))).. (-32.40 = -32.64 + 0.24)

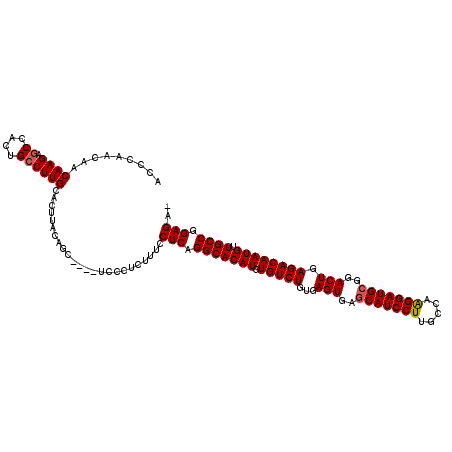
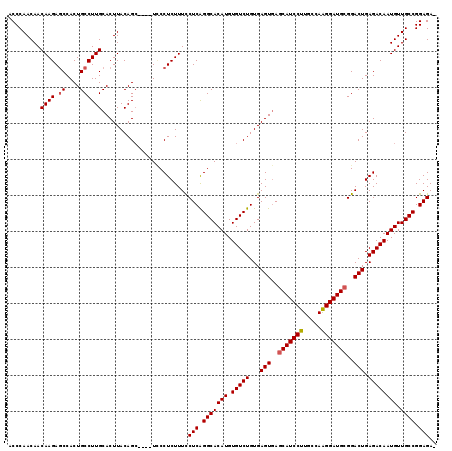
| Location | 21,906,298 – 21,906,412 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 94.91 |
| Mean single sequence MFE | -43.22 |
| Consensus MFE | -36.56 |
| Energy contribution | -36.80 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21906298 114 - 22407834 -UCUCCGGCAACAUUGUCUCAGUCCGCAUCCUUGGCAAGGAUGCUCACUCACAGACACAUGUGCCUGAGAAAAGAGGGA----GCUGUAAGUGCAAGGCAGUGGCUCUUGUUGUUGGGU -..(((((((((....((((((...(((((((.....))))))).....((((......)))).)))))).....((((----((..(...(((...))))..))))))))))))))). ( -42.50) >DroSec_CAF1 194934 118 - 1 -UCUCCGGCAACAUUGUCUCAGUCCGCAUCCUUGGCAAGGAUGCUCACUCACAGACACAUGUGCCUGAGGAAAGAGGCAUGUGCCUGUAAGUGCAAGGCAGUGGCUCUUGUUGUUGGGU -..((((((((((..(((.(.(((.(((((((.....))))))).((((.((((.(((((((..((......))..))))))).)))).))))...))).).)))...)))))))))). ( -49.50) >DroSim_CAF1 138965 114 - 1 -UCUCCGGCAACAUUGUCUCAGUCCGCAUCCCUGGCAAGGAUGCUCACUCACAGACACAUGUGCCUGAGGAAAGAGGGA----GCUGUAAGUGCAAGGCAGUGGCUCUUGUUGUUGGGU -..((((((((((....(((..(((((((((.......))))))(((..((((......))))..))))))..)))(((----((..(...(((...))))..))))))))))))))). ( -43.50) >DroEre_CAF1 158502 115 - 1 GUCUCCGGCAACAUUGUCUCAGUCCGCAUCCUUGGCAAGGAUGUUCACUCACAGACACAUGUGCCCGAGGAAAGAGGGA----GCUGUAAGUGCAAGGCAGUGGCUCUUGUUGUUGGGU ...((((((((((....(((..((((((((((.....)))))))((...((((......))))...)))))..)))(((----((..(...(((...))))..))))))))))))))). ( -40.30) >DroYak_CAF1 155088 114 - 1 -UCUCCGGCAACAUUGUCUCAGUCCGCAUCCUUGGCAAGGAUGCUUACUCACAGACACAUGUGCCUGAGGAAAGAGGAA----GCUGUAAGUGCAAGGCAGUGGUUCUUGUUGUUGGGA -..(((((((((....((((((...(((((((.....))))))).....((((......)))).))))))...(((((.----(((((.........)))))..)))))))))))))). ( -40.30) >consensus _UCUCCGGCAACAUUGUCUCAGUCCGCAUCCUUGGCAAGGAUGCUCACUCACAGACACAUGUGCCUGAGGAAAGAGGGA____GCUGUAAGUGCAAGGCAGUGGCUCUUGUUGUUGGGU ...(((((((((....((((((...(((((((.....))))))).....((((......)))).))))))...(((((.....(((((.........)))))..)))))))))))))). (-36.56 = -36.80 + 0.24)

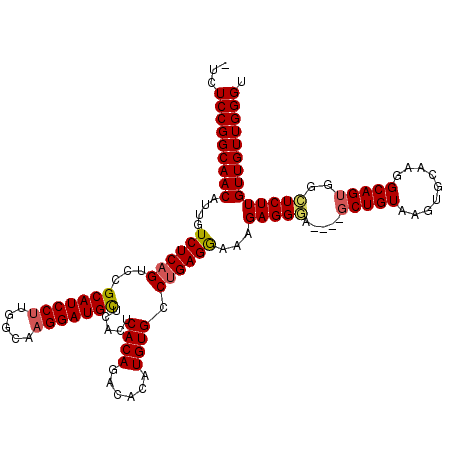
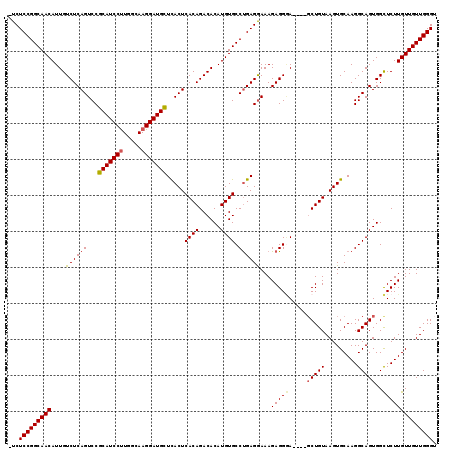
| Location | 21,906,334 – 21,906,440 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -34.94 |
| Energy contribution | -34.98 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21906334 106 + 22407834 -UCCCUCUUUUCUCAGGCACAUGUGUCUGUGAGUGAGCAUCCUUGCCAAGGAUGCGGACUGAGACAAUGUUGCCGGAGA------CGGAGAAGGAUACUGCCACGAAGACAAA -(((.(((((((((.(((((((.(((((...(((..(((((((.....)))))))..))).)))))))).)))).))))------.))))).))).................. ( -39.50) >DroSec_CAF1 194973 107 + 1 AUGCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUGAGCAUCCUUGCCAAGGAUGCGGACUGAGACAAUGUUGCCGGAGA------CGGAGAAGGAUACUGCCACGAAGACAAA ...(((.(((((...(((((((.(((((...(((..(((((((.....)))))))..))).)))))))).))))(....------)))))))))................... ( -37.20) >DroSim_CAF1 139001 106 + 1 -UCCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUGAGCAUCCUUGCCAGGGAUGCGGACUGAGACAAUGUUGCCGGAGA------CGGAGAAGGAUACUGCCACGAAGACAAA -..(((.(((((...(((((((.(((((...(((..(((((((.....)))))))..))).)))))))).))))(....------)))))))))................... ( -37.60) >DroEre_CAF1 158538 112 + 1 -UCCCUCUUUCCUCGGGCACAUGUGUCUGUGAGUGAACAUCCUUGCCAAGGAUGCGGACUGAGACAAUGUUGCCGGAGACGGAGACGGAGAAGGAUACUGCCACGAAGACAAA -(((.((((..(((.(((((((.(((((...(((...((((((.....))))))...))).)))))))).)))).))).((....)))))).))).................. ( -36.70) >DroYak_CAF1 155124 106 + 1 -UUCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUAAGCAUCCUUGCCAAGGAUGCGGACUGAGACAAUGUUGCCGGAGA------CGGAGAAGGAUACUGCCAAGAAGACAAA -.((((.(((((...(((((((.(((((...(((..(((((((.....)))))))..))).)))))))).))))(....------)))))))))).................. ( -39.00) >consensus _UCCCUCUUUCCUCAGGCACAUGUGUCUGUGAGUGAGCAUCCUUGCCAAGGAUGCGGACUGAGACAAUGUUGCCGGAGA______CGGAGAAGGAUACUGCCACGAAGACAAA .....(((((.(((.(((((((.(((((...(((..(((((((.....)))))))..))).)))))))).)))).)))........((...((....)).))..))))).... (-34.94 = -34.98 + 0.04)

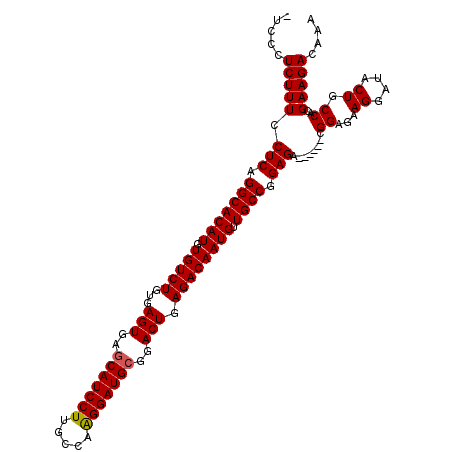
| Location | 21,906,334 – 21,906,440 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -35.98 |
| Energy contribution | -36.26 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.60 |
| SVM RNA-class probability | 0.999926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21906334 106 - 22407834 UUUGUCUUCGUGGCAGUAUCCUUCUCCG------UCUCCGGCAACAUUGUCUCAGUCCGCAUCCUUGGCAAGGAUGCUCACUCACAGACACAUGUGCCUGAGAAAAGAGGGA- .(((((.....)))))..(((((((...------((((.((((.((((((((.(((..(((((((.....)))))))..)))...))))).))))))).))))..)))))))- ( -43.50) >DroSec_CAF1 194973 107 - 1 UUUGUCUUCGUGGCAGUAUCCUUCUCCG------UCUCCGGCAACAUUGUCUCAGUCCGCAUCCUUGGCAAGGAUGCUCACUCACAGACACAUGUGCCUGAGGAAAGAGGCAU .(((((.....)))))....(((((...------((((.((((.((((((((.(((..(((((((.....)))))))..)))...))))).))))))).))))..)))))... ( -37.80) >DroSim_CAF1 139001 106 - 1 UUUGUCUUCGUGGCAGUAUCCUUCUCCG------UCUCCGGCAACAUUGUCUCAGUCCGCAUCCCUGGCAAGGAUGCUCACUCACAGACACAUGUGCCUGAGGAAAGAGGGA- .(((((.....)))))..(((((((...------((((.((((.((((((((.(((..((((((.......))))))..)))...))))).))))))).))))..)))))))- ( -40.90) >DroEre_CAF1 158538 112 - 1 UUUGUCUUCGUGGCAGUAUCCUUCUCCGUCUCCGUCUCCGGCAACAUUGUCUCAGUCCGCAUCCUUGGCAAGGAUGUUCACUCACAGACACAUGUGCCCGAGGAAAGAGGGA- .(((((.....)))))........(((.(((...((((.((((.((((((((.(((..(((((((.....)))))))..)))...))))).))))))).))))..))).)))- ( -39.10) >DroYak_CAF1 155124 106 - 1 UUUGUCUUCUUGGCAGUAUCCUUCUCCG------UCUCCGGCAACAUUGUCUCAGUCCGCAUCCUUGGCAAGGAUGCUUACUCACAGACACAUGUGCCUGAGGAAAGAGGAA- .(((((.....)))))..((((.((...------((((.((((.((((((((.(((..(((((((.....)))))))..)))...))))).))))))).))))..)))))).- ( -39.30) >consensus UUUGUCUUCGUGGCAGUAUCCUUCUCCG______UCUCCGGCAACAUUGUCUCAGUCCGCAUCCUUGGCAAGGAUGCUCACUCACAGACACAUGUGCCUGAGGAAAGAGGGA_ .(((((.....)))))..(((((((.........((((.((((.((((((((.(((..(((((((.....)))))))..)))...))))).))))))).))))..))))))). (-35.98 = -36.26 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:27 2006