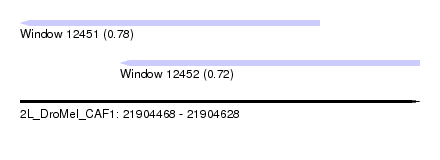
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 21,904,468 – 21,904,628 |
| Length | 160 |
| Max. P | 0.778857 |

| Location | 21,904,468 – 21,904,588 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -28.79 |
| Energy contribution | -28.27 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21904468 120 - 22407834 CUGACUUAUGGCAAUGGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUUACUACUCCAGUUGGGUAACUUCCCAGCUUAUCGUGCGUACAGUCACACACAAAAAUGGGUGAAGCCAUUGG ..........(((((((((....)))))))))....(((((((..(((.(((((...(((((((......)))))))....))).))))).)))))))....(((((......))))).. ( -39.10) >DroSec_CAF1 193133 120 - 1 CUGACAUAUGGCAAUGGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUUACUAUCCCAGUUGGGUAACUUCCCAGCUUAUCGUGCGUACAGUCACACACAAAAGUGGGUGAAGCCAGUGG (((((.(((((((((((((....)))))))))((.(((.((((....))))))).))(((((((......)))))))...)))).)).)))((((.(((....))).))))......... ( -38.30) >DroSim_CAF1 137164 120 - 1 CUGACAUAUGGCAAUGGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUUACUAUCCCAGUUGGGUAACUUCCCAGCUUAUCGUGCGUACAGUCACACACAAAAGUGGGUGAAGCCAGUGG (((((.(((((((((((((....)))))))))((.(((.((((....))))))).))(((((((......)))))))...)))).)).)))((((.(((....))).))))......... ( -38.30) >DroEre_CAF1 156736 120 - 1 UCGACAUAUGGCAAUGGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUCACAACCCCAGUUGGGUAACUACCCAGCACAUCGUGAGUACACUCGCAAACAAAAGUGGGCGAGGCCAGUGG (((((((.(.(((((((((....))))))))).).))))((((....))))...(((((((((((....))))))).....(((((....))))).........)))))))......... ( -43.80) >DroYak_CAF1 153303 120 - 1 CCGACAUAUGGCAAUAGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUUAUAACCCCAGUUGGGUAACUUCCCAGCACAUCGUGGGUACACUCGCAAACAAAAGUGGGUGAAGCCAGUGG ...((((((.(((((((((....)))))))))...))))))..............(((((((((......))))))......(((.(.(((((((........))))))).).))).))) ( -42.60) >DroAna_CAF1 135694 114 - 1 CUCGUACUUUUCGAUGAUCUCCUCGUUUUUGGUGAAAGAAUGCAGGGUUAUGACCCCAGUUUGUUCAUUUCCCAGCUUUUCGUGGGUGAUGUCGUACACAAAAACCGGGAAAGU------ .(((.......))).....((((.((((((((((...((.....((((....)))).........((((.((((........)))).)))))).))).))))))).))))....------ ( -27.50) >consensus CUGACAUAUGGCAAUGGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUUACUACCCCAGUUGGGUAACUUCCCAGCUUAUCGUGCGUACAGUCACACACAAAAGUGGGUGAAGCCAGUGG ...((((((.(((((((((....)))))))))...)))))).............((((((((((......)))))).....(((.((......)).))).....))))............ (-28.79 = -28.27 + -0.52)



| Location | 21,904,508 – 21,904,628 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -28.39 |
| Energy contribution | -27.07 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 21904508 120 - 22407834 CGUUCCAAAUGGAGACGGAUGCGAAGGCCAACGAUUCGCACUGACUUAUGGCAAUGGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUUACUACUCCAGUUGGGUAACUUCCCAGCUUAU .........(((((......((....)).((((.(((((((...((....(((((((((....)))))))))))...))))))).))))....)))))((((((......)))))).... ( -42.20) >DroSec_CAF1 193173 120 - 1 CGUUCCAAAUGGAGACGGAUGCAAAGGCCAGUGAUUCGCACUGACAUAUGGCAAUGGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUUACUAUCCCAGUUGGGUAACUUCCCAGCUUAU .(((((....))).))((((((....)).((((((...(((..((((.(.(((((((((....))))))))).).)))))))....)))))))))).(((((((......)))))))... ( -40.60) >DroSim_CAF1 137204 120 - 1 CGUUCCAAAUGGAGACGGAUGCGAAGGCCAGUGAUUCGCACUGACAUAUGGCAAUGGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUUACUAUCCCAGUUGGGUAACUUCCCAGCUUAU .(((((....))).))((((((....)).((((((...(((..((((.(.(((((((((....))))))))).).)))))))....)))))))))).(((((((......)))))))... ( -43.30) >DroEre_CAF1 156776 120 - 1 CGUUCCAAAUGGAGACGGAUGCGAAGGCCAGUGAUUCCCGUCGACAUAUGGCAAUGGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUCACAACCCCAGUUGGGUAACUACCCAGCACAU ((((((..(((..(((((..((....)).(....)..)))))..)))...(((((((((....)))))))))))))))(((((....)))))......(((((((....))))))).... ( -47.20) >DroYak_CAF1 153343 120 - 1 CGUUCCAAAUGGAGACGGAUGCGAAGGCCAUUGAUUCCCCCCGACAUAUGGCAAUAGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUUAUAACCCCAGUUGGGUAACUUCCCAGCACAU ((((((....((((.(((..((....))..))).))))............(((((((((....)))))))))))))))....................((((((......)))))).... ( -37.60) >DroAna_CAF1 135728 120 - 1 CGUUCCACGAGCAGACCGACUUGAAGGACAGAGUUUCCUUCUCGUACUUUUCGAUGAUCUCCUCGUUUUUGGUGAAAGAAUGCAGGGUUAUGACCCCAGUUUGUUCAUUUCCCAGCUUUU ......(((((.(((.((....((((((.......))))))(((.......))))).))).)))))..((((.((((((((((.((((....))))..))..)))).))))))))..... ( -29.10) >consensus CGUUCCAAAUGGAGACGGAUGCGAAGGCCAGUGAUUCCCACUGACAUAUGGCAAUGGCCUCCUGGUUAUUGCGGAAUGUGUGAAAUGUUACUACCCCAGUUGGGUAACUUCCCAGCUUAU ...(((....)))((((...((....))...............((((((.(((((((((....)))))))))...))))))....)))).........((((((......)))))).... (-28.39 = -27.07 + -1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:23 2006